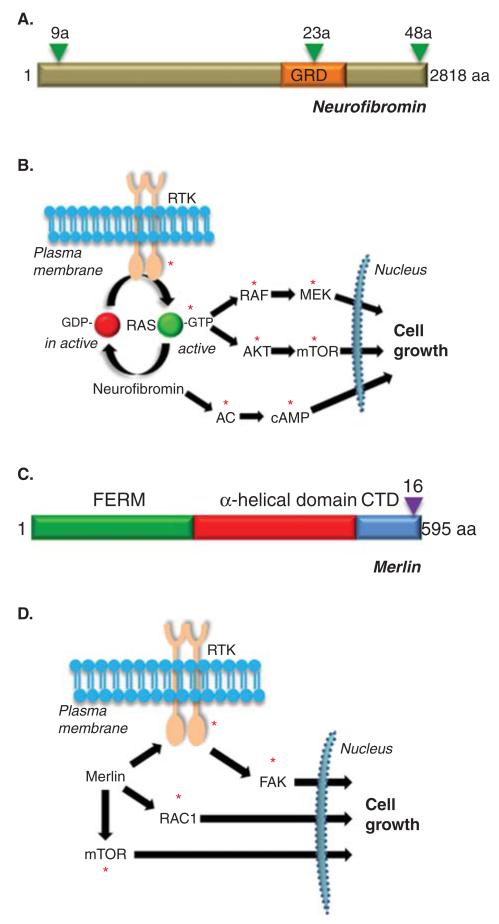
Figure 3. Structure and function of NF1 and NF2 proteins.
(A) Structure of the predicted NF1 protein, neurofibromin. GRD = GAP-related domain. Triangles denote the three known alternatively spliced exons (9a, 23a and 48a). (B) Neurofibromin functions as a negative regulator of the RAS proto-oncogene and inhibits RAS activity and downstream RAS effector (RAF/MEK and AKT/mTOR) signaling. In addition, neurofibromin positive regulates adenylyl cyclase (AC) function and cyclic AMP (cAMP) generation. (C) Structure of the predicted NF2 protein, merlin or schwannomin. FERM = protein 4.1, ezrin, radixin, moesin domain. CTD = carboxyl terminal domain. The triangle denotes the alternatively spliced exon 16. (D) Merlin negatively regulates a number of intracellular signaling molecules, including mTOR, RAC1 and FAK, depending on the cell type examined. The red asterisks denote signaling intermediates that serve as potential targets for therapeutic drug design.