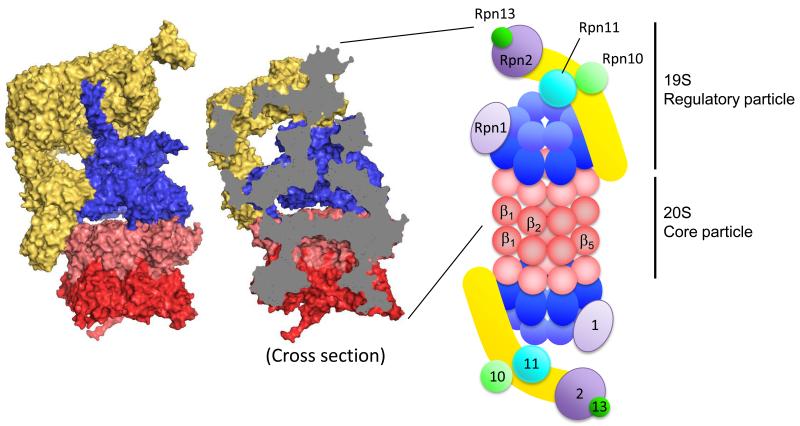
Figure 1.
Structure of the 26S proteasome. Molecular surface of the 19S activator particle bound to the 20S core particle (PDB 4C0V) (left). The 20S core particle is composed of central two β rings (dark red) and outer two α rings (light red) at either end. The 19S regulatory particle, which contains AAA ATPase subunits (blue) and non-ATPase subunits (yellow), caps either end of the 20S. Cross section reveals the degradation channel that connects the proteolytic chamber in the 20S core particle to the entrance of 19S activator (middle). Structures are produced by PyMOL. Schematic drawing of the 26S proteasome indicates the approximate locations of the enzymatic activities and binding platforms on the 19S activator cap (right). α (light red) and β (dark red) subunits of the 20S particle, ATPase domain (dark blue) and OB domain (light blue) of ATPase subunits, backbone of lid sub particle (yellow), docking subunits Rpn1 (light purple) and Rpn2 (dark purple), ubiquitin receptors Rpn10 (light green) and Rpn13 (dark green), and DUB metallo-protease subunit Rpn11 (sky blue).