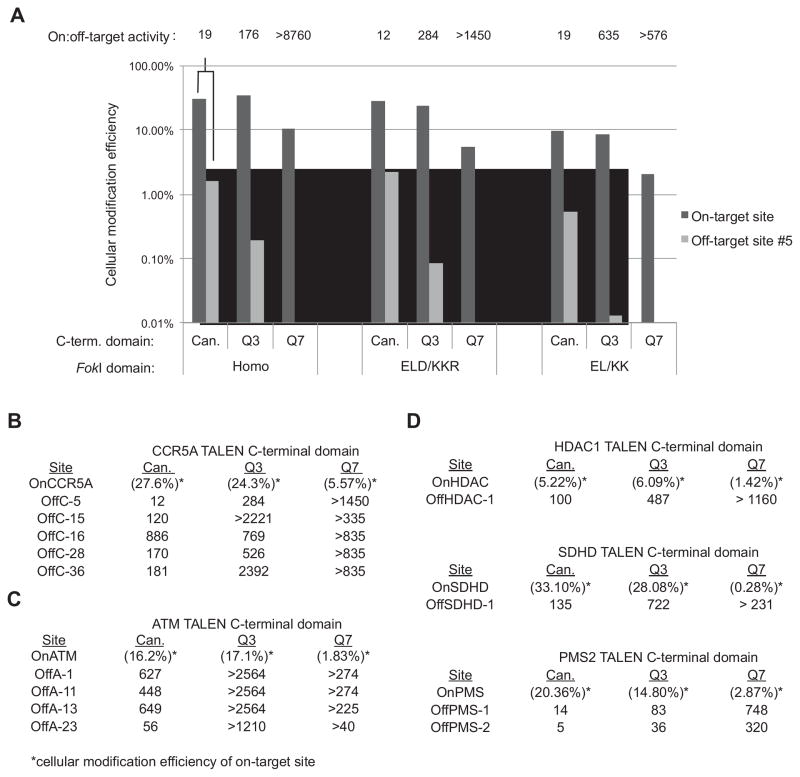
Figure 6. Specificity of engineered TALENs in human cells.
(A) The cellular modification efficiency of canonical and engineered TALENs expressed as a percentage of indels consistent with TALEN-induced modification out of total sequences is shown for the on-target CCR5A site (OnCCR5A) and for CCR5A off-target site #5 (OffC5), the most highly cleaved off-target substrate tested. All pairwise P-values comparing the number of observed sequences containing insertions or deletions consistent with TALEN-induced cleavage vs. the total number of sequences were calculated with a Fischer exact test between samples (see Supplementary Table S7). P-values are < 0.005 for samples of canonical vs. Q3 vs. Q7 TALENs in the same FokI background for both on-target and off-target sites with the exception of off-target site #5 modified with Q3 vs. Q7 TALENs in the EL/KK FokI background (P-value < 0.087). On:off target activity, defined as the ratio of on-target to off-target modification, is shown above each pair of bars. (B) The on:off target activity of the canonical, Q3, and Q7 TALENs for each detected genomic off-target substrate of the CCR5A TALEN with the ELD/KKR FokI domain are shown. The absolute genomic modification frequency for the on-target site is in parentheses. (C) Same as (B) for the ATM TALENs and off-target sites. (D) The on:off target activities of the canonical, Q3, and Q7 TALENs for each detected genomic off-target substrate of the PMS2, SDHD, and HDAC1 TALENs with the ELD/KKR FokI domain are shown. The absolute genome modification frequency for the on-target site is in parentheses.