Figure 1.
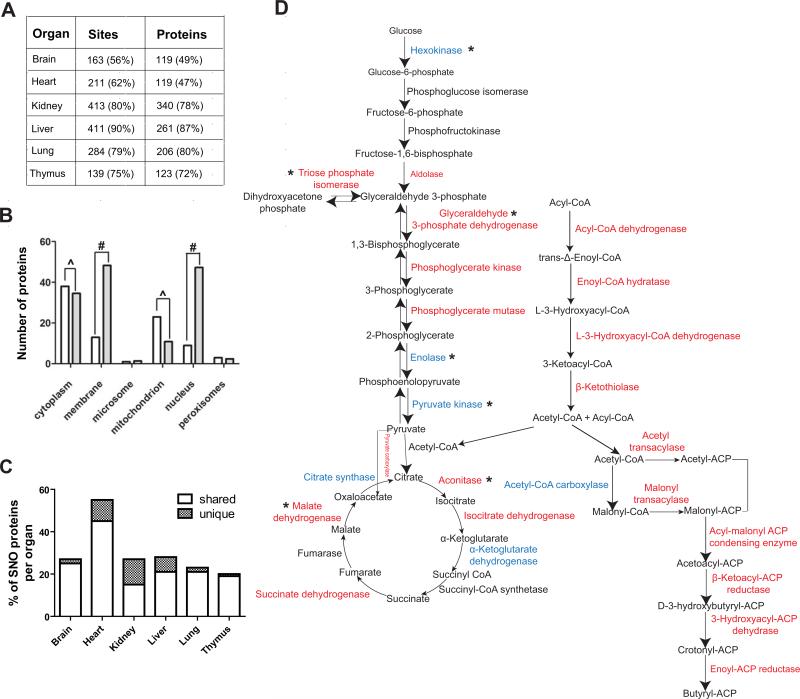
Overview of the cysteine S-nitrosoproteome of the mouse. (A) The number of sites and proteins identified across six organs in wild-type mouse and their dependency to eNOS activity as percentage of the wild type is indicated in parenthesis. Three biological replicates from each organ from wild type and eNOS−/− mice were analyzed. (B) Sub-cellular localization of the S-nitrosoproteome (white bars) and comparison with the rest of the mouse proteome (grey bars); ^ indicate overrepresentation and # underrepresentation, with both indicating p<0.0001. (C) Percentage of S-nitrosylated mitochondrial proteins. The white bar indicates proteins shared with at least another organ. The hatched bar indicates unique proteins for a particular organ. (D) S-nitrosylated enzymes present in wild-type mouse liver that were absent in eNOS−/− liver and that are involved in glucose metabolism, TCA cycle and fatty acid metabolism are marked in red. S-nitrosylated proteins that are present in eNOS−/− liver are depicted in blue. Asterisk (*) indicates proteins that have been reported to be S-nitrosylated. For all the proteins and sites identified see tables S1 to S6.