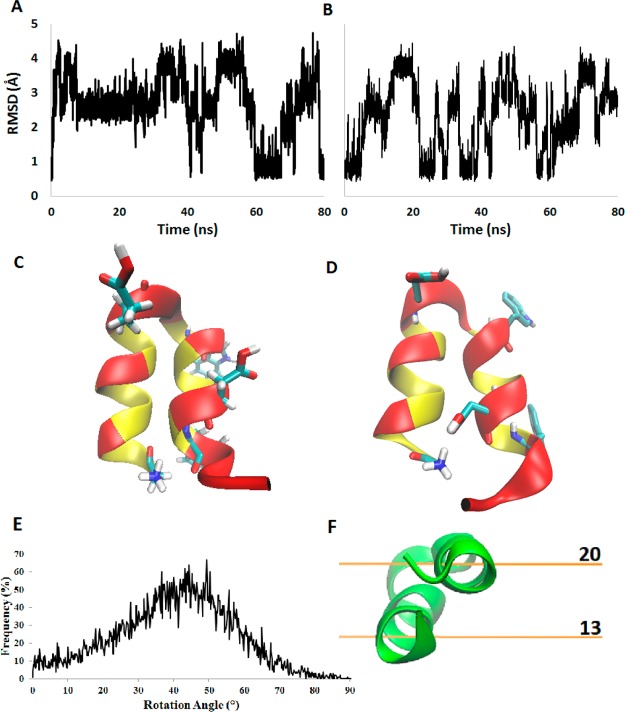
Figure 7.
IMM1 trajectory analysis. Root mean square deviations from the NMR structure over MD trajectories for backbone atoms in the HAFP hairpin. MD trajectories were simulated for 100 ns using the IMM1 implicit membrane with (A) protonated Glu/Asp and (B) unprotonated Glu/Asp. Hairpin structures resulting from (C) a 200 ns explicit simulation and (D) a 100 ns implicit simulation are displayed with acidic residues, tryptophan, and G1 residues highlighted. (E) Population of rotation angle values sampled over the low pH simulation. (F) Hairpin structure shown with relative membrane depth values (z axis).