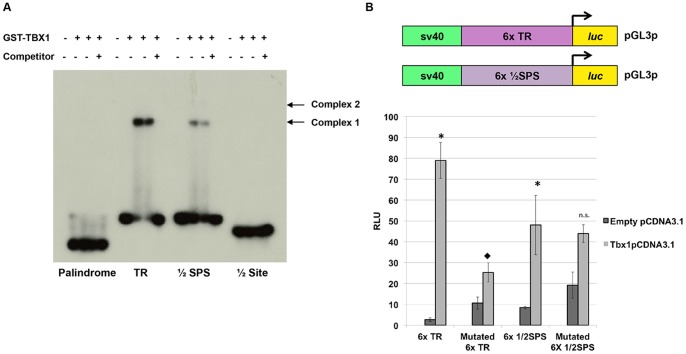
Figure 2. Specificity of new binding sites tested via EMSA and luciferase assays.
A: The binding sites identified from the SELEX experiment were tested with GST-TBX1 separately by EMSA to determine if there is specific binding (Tandem Repeat, TR; Half Site Partial Site, ½SPS, half site; ½ Site). For comparison, the palindromic T-site was also tested but binding was very weak as compared to the newly identified binding sites and was only observed after extensive overexposure of the autoradiogram (not shown). B: Luciferase reporter constructs containing 6 copies (6x) of the TR and 6x of the ½SPS, respectively, were co-transfected with full length Tbx1-pCDNA3.1 and compared to the empty pCDNA3.1 transfection to determine if TBX1 could activate transcription of a reporter via these sites. A significant increase of luciferase activity was observed in the presence of full-length TBX1 for both the 6x TR and the 6x½SPS when compared to transfection of the empty pCDNA3.1 vector (TR: 29 fold; Students t test, *p<0.001; ½SPS: 5.6 fold; Students t-test, *p<0.02). The mutations analyzed were those previously tested in a half site where AGGTGTGA was mutated to AATTTTGA [31]. When these nucleotide changes were present in the TR, there was a dramatic decrease in activation by Tbx1-pCDNA3.1 (7.4 fold; Students t test, ♦p<0.001). The same mutation in the 6x½SPS construct did not show a significant change when compared to the normal ½SPS (n.s. not significant). All data are presented as means ±SD; n≥3.