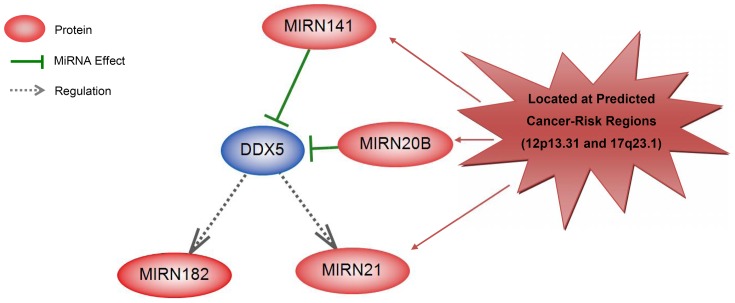
Figure 2. Subnetwork center on DDX5 derived from network of common altered variants in different cancers.
Network is including mir-21, mir-182, -mir20b and mir-141. Network was constructed using pathway studio 9 software. Network was assembled based on bioinformatics and literature, combined with biological interpretation of the microarray data and enriched Gene Ontology functional groups. Red: over-regulated entities in most of cancers. Blue: down-regulated entities in most of cancers.  represents negative-regulated.
represents negative-regulated.