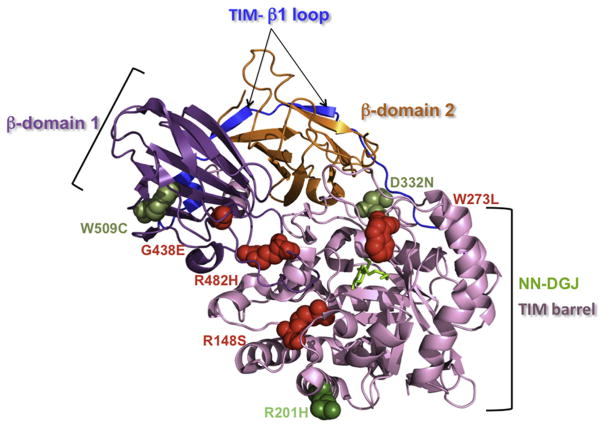
Fig. 4.
Localization of the missense mutations analyzed in this report on the crystal structure of the human β-Gal monomer: The β-Gal monomeric structure is shown in ribbon representation with the bound NN-DGJ (in green) inserted inside the active site pocket, and the mutated amino acid residues are represented as spheres within the diagram. The different domains of the β-Gal are indicated (TIM barrel domain, in pale pink; β domain 1, in purple; β domain 2, in orange and TIM- β1 loop, in blue). Our data identified p.R201H (dark green, on the surface of the protein) as a mutation that strongly responds to NN-DGJ (14771 and 3633, Table 1 and 2), while p.W273L (4080, Table 1, involved in ligand recognition), p.G438E (10213, Table 1, on the surface of the protein), p.R482H (4080 and 8981, Table 1, core residue in the interdomain), and p.R148S (8981, Table 1, core residue in the intradomain) are nonresponsive mutations (in red). Taken together these data also indicate that p.W509C (3633, Table 1, core residue in the intradomain) and p.D332N (4992, Table 1, on the surface of the protein) are responsive mutations (lighter green). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)