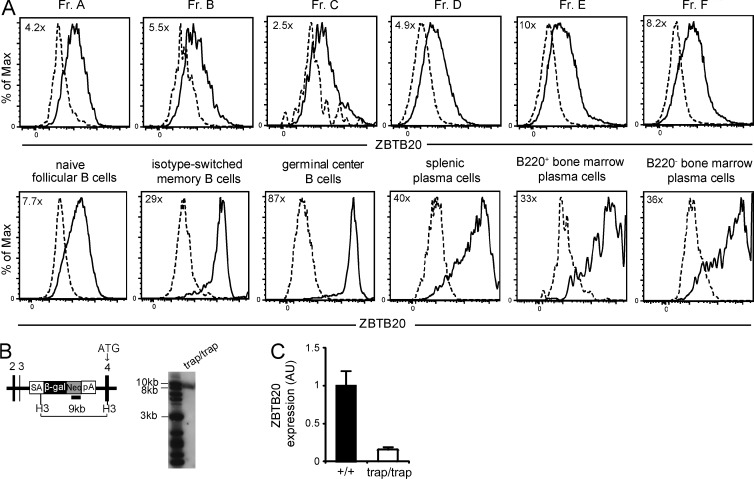
Figure 1.
ZBTB20 is highly expressed by activated B cells. (A) Flow cytometry analysis of ZBTB20 expression through β-galactosidase expression in Zbtb20+/trap B cell progenitors by the Hardy classification (Fr. A–F), naive B cells, splenic plasma cells, germinal center B cells, isotype-switched memory B cells, and B220+ and B220− BM plasma cells. Gating strategies are shown in Fig. S1. Cells from heterozygous Zbtb20+/trap (solid lines) and control Zbtb20+/+ (dashed lines) mice were both stained with the β-galactosidase substrate fluorescein di-β-galactopyranoside. Values in the top left corner of plots represent fold changes of the mean fluorescence intensities between Zbtb20+/trap and Zbtb20+/+ cells. Data are representative of four independent experiments with one mouse per group per experiment. (B) Map of Zbtb20 locus (left) and Southern blot analysis (right) demonstrating the correct location and copy number of the gene trap insertion. Black numbered rectangles denote exons 2–4 of Zbtb20. The small solid rectangle indicates the location of the Neo-specific probe. ATG, translation start site; β-gal, β-galactosidase; H3, HindIII; Neo, neomycin phosphotransferase; pA, polyadenylation site; SA, splice acceptor. (C) qRT-PCR analysis of ZBTB20 transcript levels in peripheral blood B cells from Zbtb20+/+ (n = 9) and Zbtb20trap/trap (n = 8) fetal liver chimeras. Transcript levels were measured in technical triplicates, and Zbtb20 expression was normalized to the mean Actb expression of each sample. Mean values ± SD are shown in arbitrary units (AU) relative to wild-type B cell levels. Data are representative of two independent experiments.