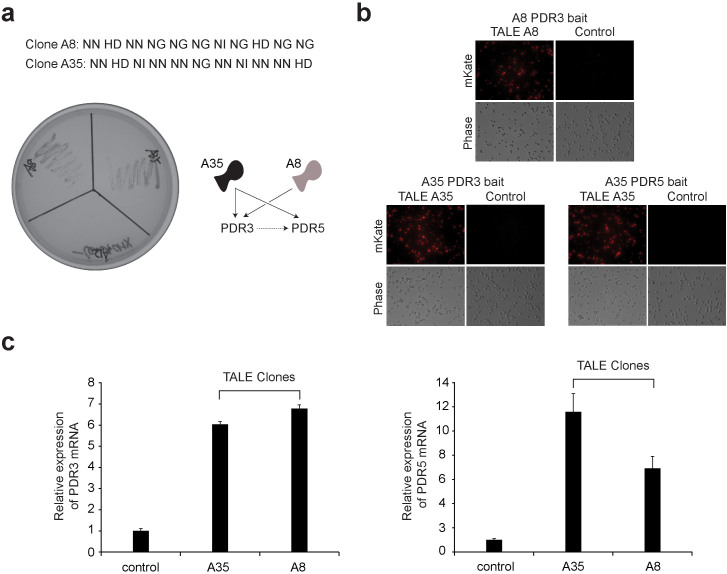
Figure 3. Genetic screen for cycloheximide resistance in yeast using the TALE-VP16 plasmid library.
(a) Confirmation of the genuine positive yeast clones conferring cycloheximide resistance. 18 positive clones were isolated from the cycloheximide resistance screening. Subsequently, these positive TALE-VP16 fusion plasmids were recovered and again re-transformed into the wild-type yeast cells. The transformed cells were then re-streaked onto –Leu plates containing 0.5 µg/ml of cycloheximide. After 3 days, cells transformed with genuine positive clones pGADT7-TALE-A8-VP16 and pGADT7-TALE-A35-VP16 were able to grow robustly. In contrast, cells transformed with the false positive clone pGADT7-TALE-A12-VP16 or the control pGADT7 failed to grow. (b) The isolated TALE-VP16 fusion clones A8 and A35 bind to the promoters of the PDR3 and PDR5 genes. TALE-VP16 fusion clones A8 and A35 were isolated from cycloheximide resistance screening. Both A8 and A35 were predicted to bind to the promoter of PDR3 gene. In addition, A35 was predicted to target the promoter of the PDR5 gene. Four copies of the predicted PDR3/PDR5 promoter targets and their immediate adjacent sequences were cloned in front of a fluorescence reporter gene (mKATE2) in yeast (bait). These yeast stable clones were then transformed with corresponding pGADT7-TALE-A8-VP16, pGADT7-TALE-A35-VP16 or pGADT7 (control). TALE-VP16 fusion clones A35 or A8 potently induced the expression of mKATE2 in yeast cells containing the corresponding baits, while pGADT7 failed to do so (right). (c) The isolated TALE-VP16 fusions A8 and A35 induced overexpression of endogenous PDR3 and PDR5 genes. Wild-type yeast cells were transformed with pGADT7-TALE-A8-VP16, pGADT7-TALE-A35-VP16 or pGADT7 (control). The expression levels of PDR3/PDR5 were measured by quantitative RT-PCR. Both clones were able to effectively induce the overexpression of PDR3 (n = 3) and PDR5 (n = 3).