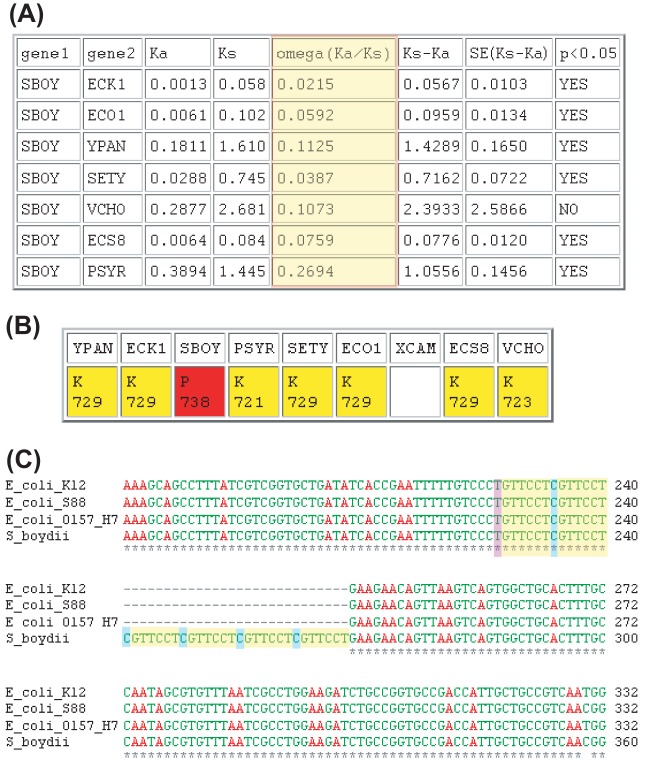
FIG 4.
Insertion in the genomic sequence of S. boydii fadB. (A) By reading through a frameshift, SearchDOGS's ORFmaker program was able to create a full-length ORF (highlighted in red) at the locus corresponding to the highly conserved fadB gene. (B) Screenshot of the SearchDOGS Ka/Ks output for the reconstructed fadB candidate against the annotated FadB orthologs. Omega (Ka/Ks) values are highlighted in red. Tests to determined 95% confidence intervals carried out on the value of Ks − Ka indicate that, in all cases except the S. boydii/V. cholerae pairwise comparison, Ks − Ka is greater than 0 with statistical significance, indicating protein sequence conservation. (C) ClustalW alignment of the nucleotide sequence surrounding an apparent expansion of a 7-bp repeat in S. boydii fadB. E. coli K-12, E. coli S88, E. coli O157:H7, and S. boydii are shown, and the repeat sequence is highlighted (83). The entire length of the insertion is 28 bp and thus causes a frameshift at this location in S. boydii fadB.