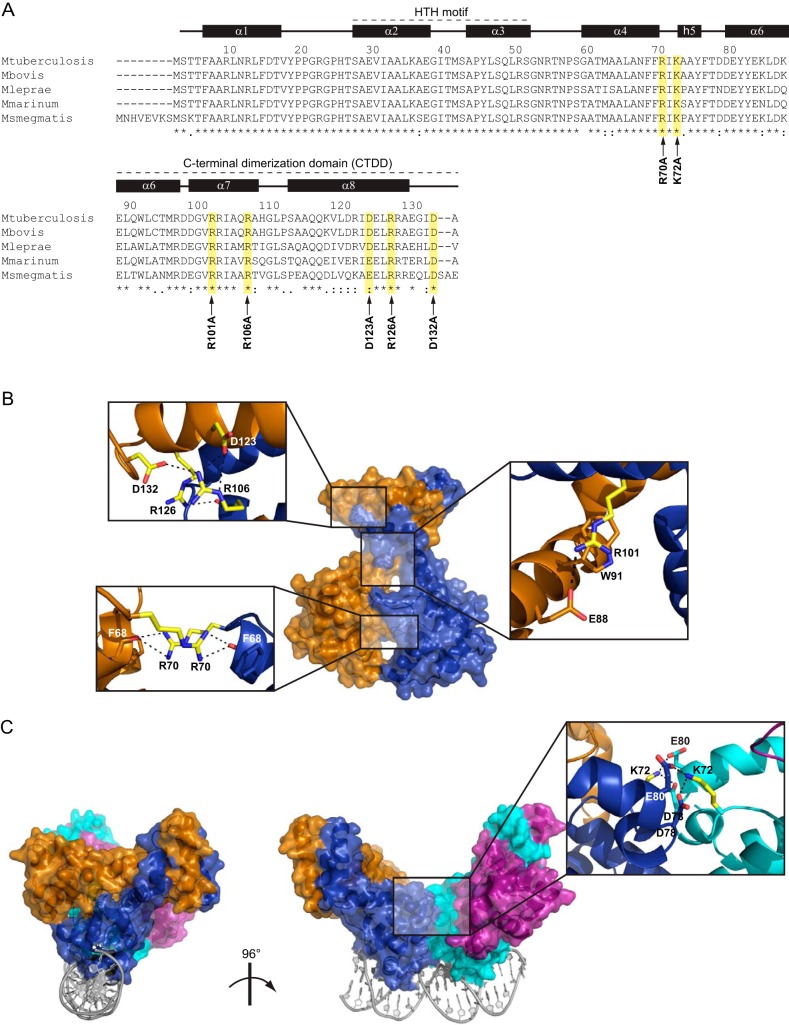
FIG 1.
Structural mapping of the amino acids replaced by Ala in EspR. (A) Sequence alignment of EspR orthologs in different mycobacteria. Amino acid positions and secondary structure referring to the M. tuberculosis H37Rv EspR crystal structure (PDB 3QF3) are specified on the top. Amino acids selected for site-directed mutagenesis are highlighted in yellow, and the corresponding point mutation is indicated with an arrow at the bottom. The N-terminal helix-turn-helix DNA-binding domain and the C-terminal dimerization domain (CTDD) containing most of residues involved in intersubunit interactions are indicated. (B) Mapping of the residues replaced by Ala (yellow) onto the EspR dimer. Interactions involving these residues in the wt structure (PDB 3QF3) are shown in magnified insets. (C) Mapping of the Ala-replaced Lys72 (yellow) onto the EspR dimer-of-dimers bound to DNA as previously obtained by X-ray crystallography (PDB 3QYX) and molecular modeling (5). Interactions involving Lys72 at the dimer-of-dimers interface (blue-cyan) are shown in the magnified inset.