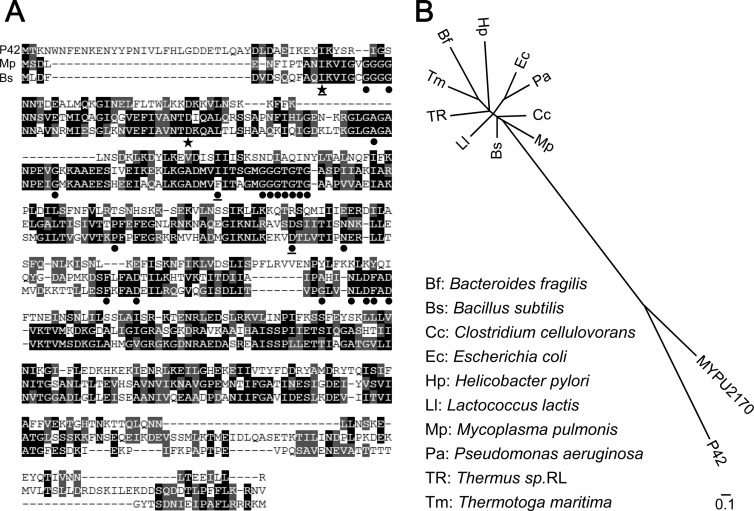
FIG 5.
Sequence analysis of P42. (A) Similarity between P42 (top rows of sequence) and FtsZs of Mycoplasma pulmonis (Mp; middle rows) and Bacillus subtilis (Bs; bottom rows). Identical and similar amino acids are marked by black and gray boxes, respectively. Twenty-four amino acids that were well conserved in FtsZ and tubulin are marked by filled circles and stars (59); those marked by a solid star were also conserved in P42. Two amino acids were not conserved in FtsZ of Mycoplasma pulmonis. Twenty-one amino acids were involved in GTP binding, and the functions of the other three amino acids (underlined) are unknown. (B) Phylogenetic tree analyzed for P42, the ortholog of M. pulmonis (MYPU2170), and FtsZ proteins from 10 species. The ftsZ gene has not been identified in the genome of M. mobile. The scale bar indicates the number of substitutions by position.