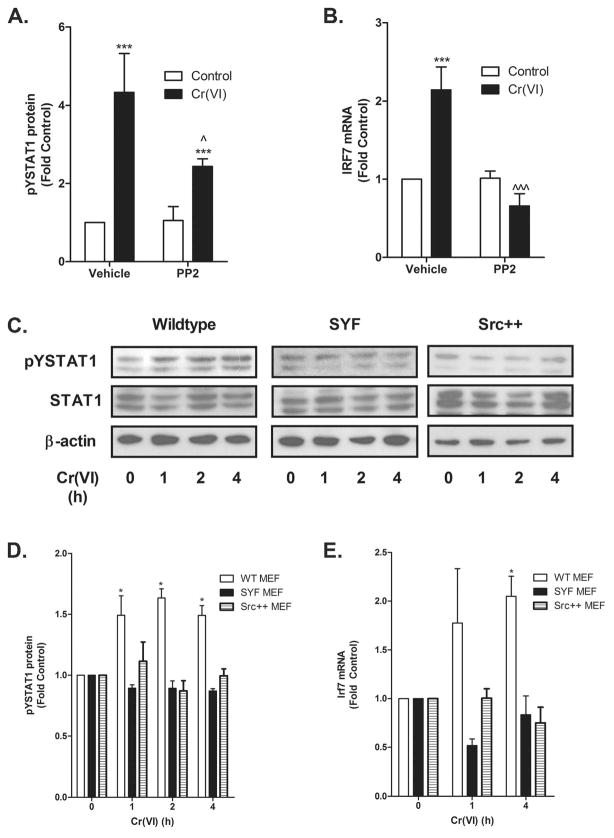
Figure 5.
Cr(VI) activation of STAT1 requires Src family kinases. A. BEAS-2B cells were pretreated with PP2 for 30 min prior to the addition of 5 μM Cr(VI) for 1 h. pYSTAT1, STAT1, and β-actin levels were determined by western analysis. Density of the protein bands from three separate experiments were quantified with ImageJ software. B. BEAS-2B cells were pretreated with PP2 for 30 min prior to the addition of 5 μM Cr(VI) for 4 h. Total RNA was isolated and IRF7 mRNA levels were measured by RT-PCR and normalized against the housekeeping gene, RPL13A. C. Wild-type (WT), SYF, and Src++ MEF cells were incubated with 5 μM Cr(VI) for the indicated times and nuclear protein was isolated. pYSTAT1, total STAT1, and β-actin levels were determined by western analysis and a representative blot from a single experiment is shown. D. Density of the protein bands from three separate experiments were quantified with ImageJ software. E. WT, SYF, and Src++ MEF cells were exposed to 5 μM Cr(VI) for the indicated times and total RNA was isolated. Irf7 mRNA levels were measured by RT-PCR and normalized to the housekeeping gene, β-actin. Data is presented as mean ± SEM of fold control (n=3). * and *** designate p<0.05 and p<0.001, respectively, compared to untreated (control) cells. ^ and ^^^ designate p<0.05 and p<0.001, respectively, compared to cells treated with Cr(VI) alone.