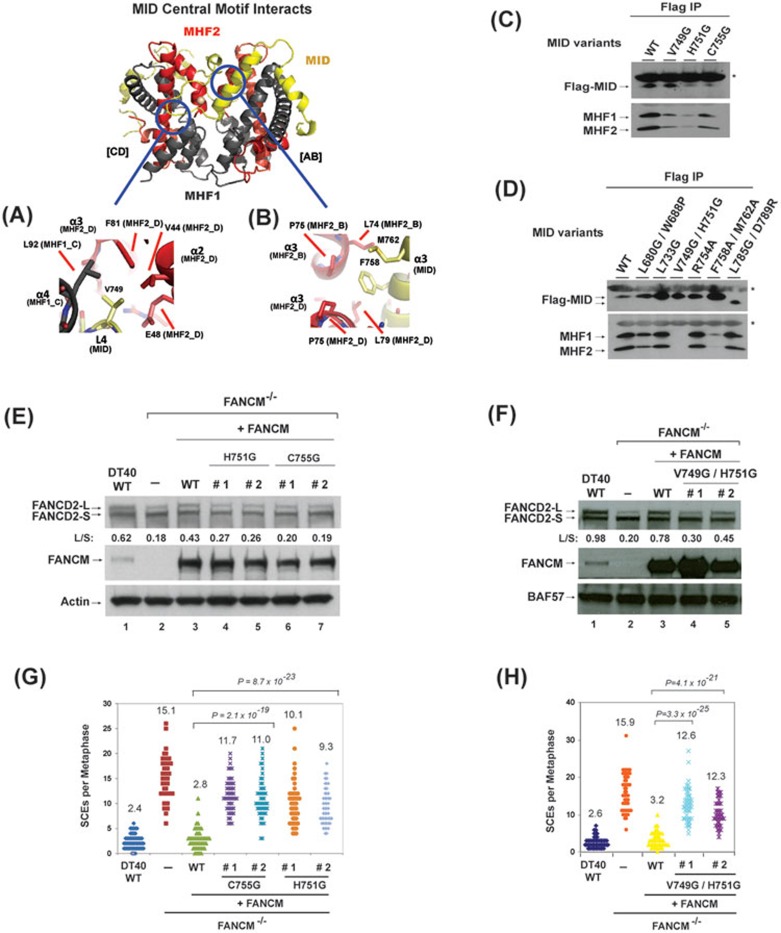
Figure 2.
Identification of the residues in MID that are essential for stabilization of the MID-MHF complex, normal activation of the FA network and suppression of SCEs. (A, B) Key MID and MHF interfaces are shown in detail. MID central motif residues: (A) L4 (MID-V749), (B) α3 amphipathic helix residues (MID-F758, M762). Proteins colored as in Figure 1A. (C, D) Co-immunoprecipitation analyses in HEK293 cells showing the association between MHF and MID variants as indicated. Asterisks indicate cross-reactive non-specific polypeptides (IgG light chains). Because the MID domain is the only domain in FANCM that binds MHF14, a mutation that disrupts MID-MHF association should have the same effect in the context of the full-length protein. (E, F) Immunoblotting shows the levels of FANCD2 and FANCM in whole-cell lysates from various DT40 cells as indicated, which include: wild-type (WT), FANCM−/− cells, and FANCM−/− cells complemented with human WT or mutants of FANCM. The ratio between the monoubiquitinated and non-ubiquitinated FANCD2 (L/S) is shown. Cells were treated with MMC (50 ng/ml) for 18 h. BAF57 or actin was used as a loading control. (G, H) Histograms showing spontaneous SCE levels of various DT40 cells as indicated. The mean number of SCEs per metaphase is listed. P-values were calculated using the Student's t-test.