Figure 4.
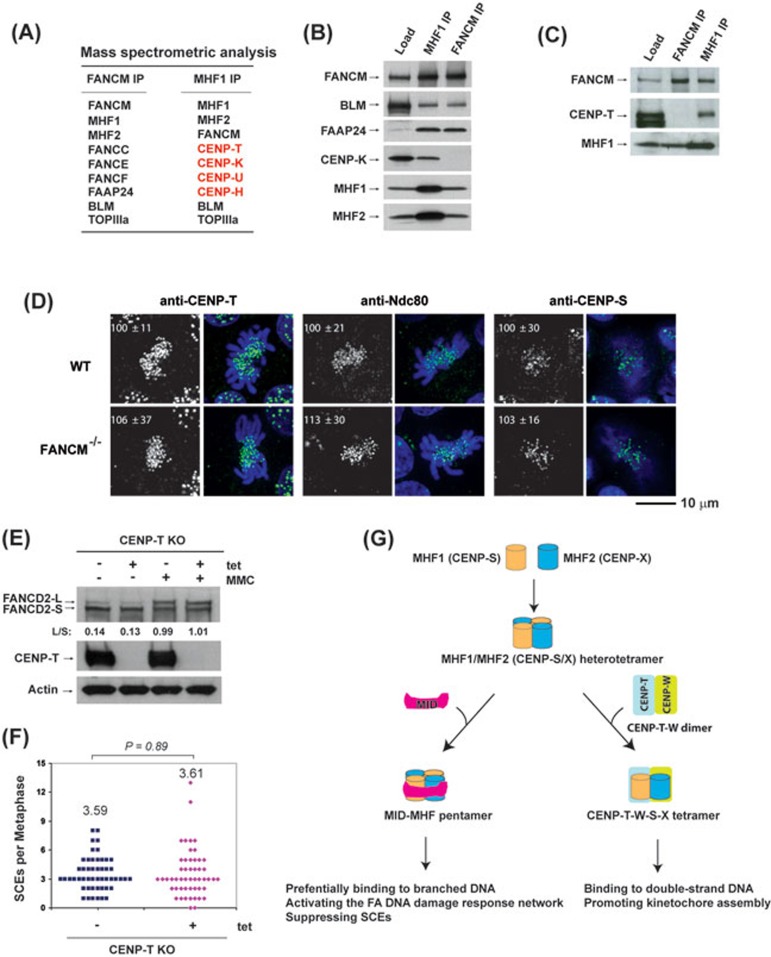
MHF1 and MHF2 are present in two functionally distinct complexes. (A) Mass spectrometric analysis shows that MHF1 (CENP-S) and MHF2 (CENP-X) are present in both FANCM and MHF1 immunoprecipitates isolated from the chromatin fraction of HeLa cells by either FANCM or MHF1 antibody, whereas other centromere proteins (CENP-T, CENP-K, CENP-U, CENP-H) were detected only in the MHF1 immunoprecipitate. (B, C) Immunoblotting shows that MHF1, MHF2, FANCM and other components of the FA core and Bloom complexes were present in both FANCM and MHF1 immunoprecipitates, but CENP-K and CENP-T were detected only in the MHF1 immunoprecipitate. (D) Immunofluorescence analyses of wild-type (WT) or FANCM−/− DT40 cells with different antibodies (green) as indicated. DNA was stained by DAPI (blue). The signal intensities of CENP-T, Ndc80 and CENP-S at each kinetochore detected by immunofluorescence were measured relative to an adjacent background signal (n > 10 cells; average intensity ± SD). The scale bar represents 10 μm. (E) Immunoblotting shows the levels of FANCD2 and CENP-T in CENP-T conditional knockout (KO) DT40 cells with or without the treatment of tetracycline (tet) for 24 h. The ratio between the monoubiquitinated and non-ubiquitinated FANCD2 (L/S) is shown. Cells were treated with MMC (50 ng/ml) for 16 h. Actin was used as a loading control. (F) Histograms showing the levels of spontaneous SCEs in CENP-T conditional KO DT40 cells with or without the treatment of tet for 24 h. The mean number of SCEs per metaphase is listed. P-value was calculated using the Student's t-test. (G) A diagram describes that MHF has two distinct functions by forming independent complexes with FANCM and CENP-T/W. One is the FANCM-MHF complex that preferentially binds branched DNA and functions specifically in DNA repair. The other is CENP-T-W-S-X complex that promotes kinetochore assembly at centromeres20.