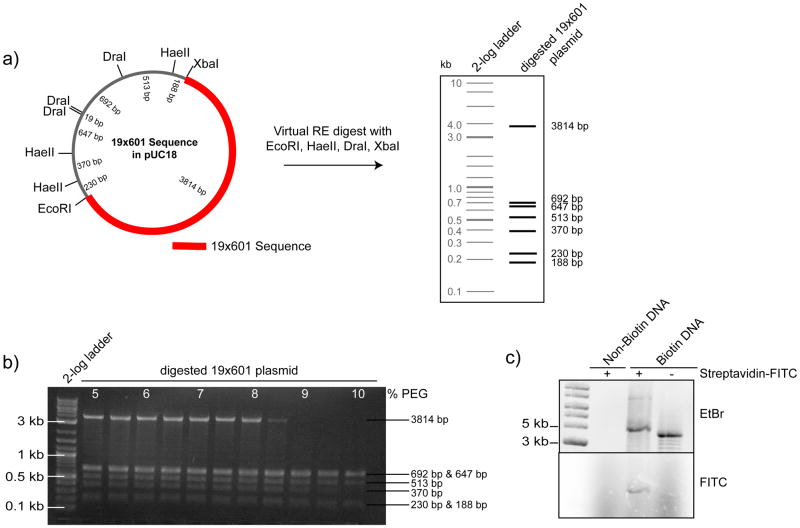
Figure 3. Generation and analysis of biotinylated DNA arrays for chromatin reconstitution.
(a) Schematic map of the 19×601 nucleosome positioning sequence plasmid (left panel) indicating the position of the 601 sequence repeats and restriction sites (RE) of interest. Result of a virtual RE digest with EcoRI, HaeII, DraI and XbaI (right panel) to facilitate the purification of the 19×601 sequence (~3800bp) from the digest plasmid backbone (fragments < 700bp) by PEG precipitation. (b) Representative agarose gel of the PEG precipitation procedure. At ~ 8.5–9% PEG the fragment of interest precipitates while DNA fragments <700bp remain soluble in the supernatant. (c) To asses the biotinylation efficiency of the array DNA (Biotin-DNA), and of non-biotinylated DNA as a control (Non-Biotin DNA), the DNA was incubated for 15 min with (+) or without (−) Streptavidin-FITC. Subsequently, the reactions were separated by agarose gel electrophoreses and the DNA was visualized by Ethidium Bromide (EtBr) stain. Bound Streptavidin-FITC (FITC) caused a gel mobility-shift and is detected by fluorescence imaging (lower panel). Figure is adapted from 2.