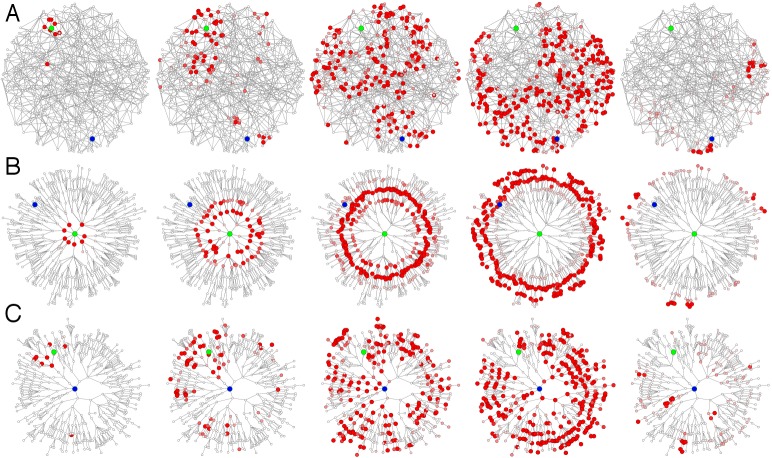
Effective distance and outbreak origin reconstruction in multi-scale network contagion processes.
(A) Each panel depicts a temporal snapshot (from left to right at equidistant time intervals) in a simple contagion process in which infected nodes (red) deliver the infection to connected nodes at a fixed rate before they recover at a another rate (SIR dynamics 38). The network consists of 512 nodes on a quasi-triangular, random lattice. Each node is connected to its nearest local neighbors. In addition to the local lattice structure, 128 long range links exist between randomly chosen pairs of nodes. The origin of the outbreak is marked in green. Because of long range connectivity the pattern quickly loses spatial structure and becomes chaotic such that it is difficult to predict from metric cues alone when the contagion arrives at a given node. More importantly, long range connectivity leads to a loss of spatial coherence and it becomes impossible to determine the origin of outbreak. (B) The same pattern as in (A) is shown in the effective distance perspective from the outbreak origin. The depicted tree is the shortest path tree, i.e. the most probable spreading path of the contagion process. Radial distance is proportional to effective distance as defined in the text. In this alternative representation the complex pattern in the conventional view is mapped onto a simple propagating wave front and arrival times are easily computed. (C) The regularity of the pattern is only present from the perspective of the actual outbreak origin. When the contagion process is viewed from any other node (here the node depicted in blue), the pattern lacks regularity.