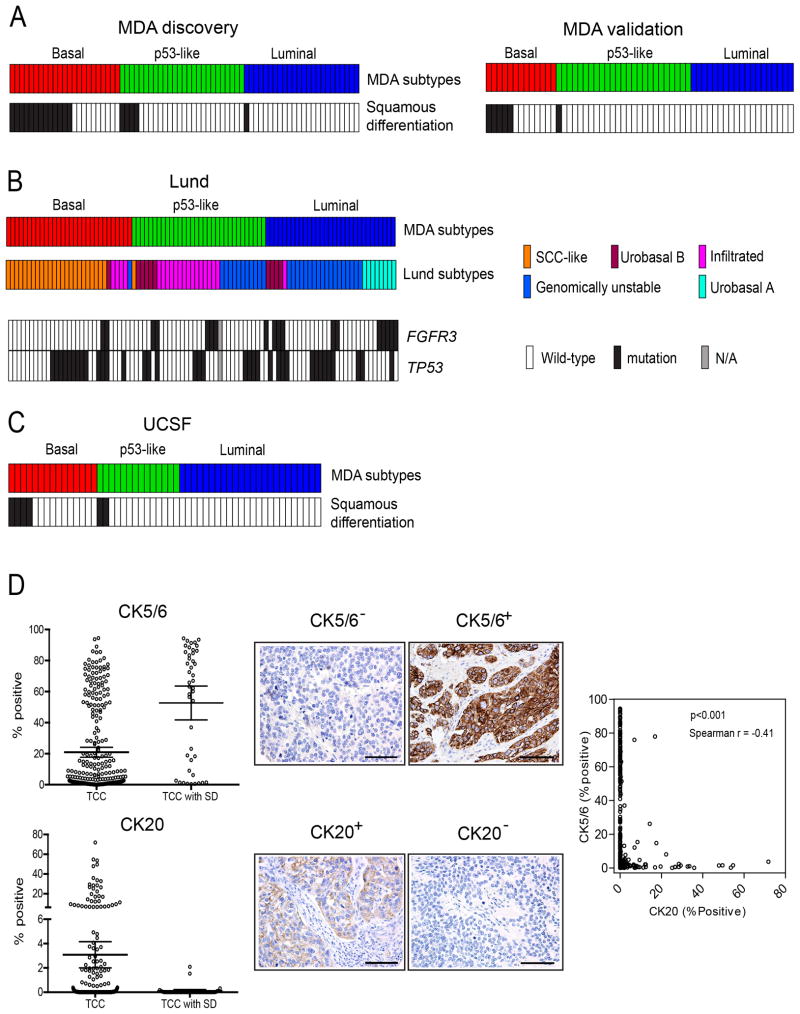
Figure 3. Presence of squamous features in the subtypes.
A. Tumor squamous feature content in the MD Anderson discovery and validation cohorts. Subtype designations are indicated by the top color bars, and the presence of squamous features (in black) is indicated in the color bars below. B. Relationship between the MD Anderson subtypes and the molecular taxonomy developed by Sjödahl et al (Sjodahl et al., 2012). Whole genome mRNA expression (Illumina platform) and clinical data were downloaded from GEO (GSE32894), and the oneNN classifier was used to assign the Lund tumors to subtypes. Subtype membership is indicated by the top color bars, and FGFR3 and TP53 mutations in the Lund tumors are indicated in color bars below. Black: mutant; white: wild-type; grey (N/A): mutation data were not available. C. Presence of squamous features in the UCSF dataset. Whole genome mRNA expression profiling (in-house platform) and clinical data were downloaded from GEO (GSE1827), and the oneNN classifier was used to assign the UCSF tumors to the subtypes. Subtype memberships for each tumor are indicated in the top color bars, and the presence of squamous features (in black) is indicated in the color bar below. D. Tissue microarray analysis of CK5/6 (basal) and CK20 (luminal) cytokeratin expression. Cytokeratin protein expression was measured by immunohistochemistry and optical image analysis in the MD Anderson Pathology Core on a tissue microarray containing 332 high-grade pT3 tumors. The percentages of positive tumor cells as determined by image analysis are shown. Left panels: mean levels of CK5/6 (top) and CK20 (bottom) in tumors without (TCC) or with (TCC with SD) squamous features. Bars indicate mean values with 95% confidence intervals. Middle panels: representative images of stained cores from tumors that expressed high or low levels of CK5/6 or CK20. The scale bars correspond to 100 microns. Right panel: relationship between CK5/6 and CK20 expression across the cohort. See also Tables S3–5 and Fig. S2.