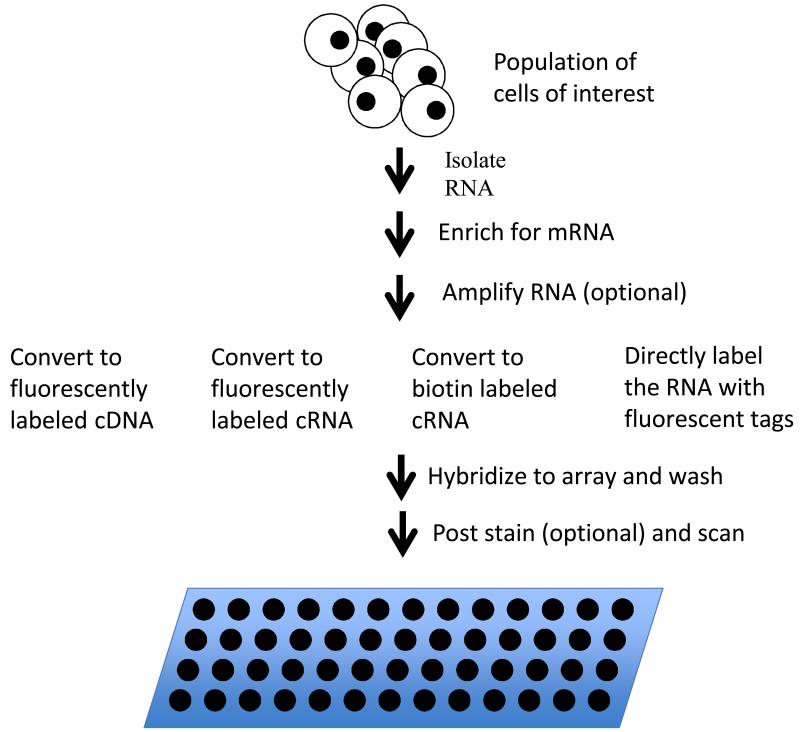
Figure 3.
Gene expression analysis via microarrays. RNA is isolated from the sample of interest and enriched for messenger RNA. In eucaryotes, poly-A tailed mRNA’s are typically enriched using affinity purification with oligo dT beads or columns. In procaryotes, unselected RNA is typically depleted for ribosomal sequences using bead or columns coated with sequences complementary to 16s. After message enriched RNA is in hand, it is optionally amplified and labeled by any one of a number of methods and the resulting labeled sample is hybridized to a microarray. The array is washed to remove unbound sample. If the sample was labeled with biotin, the array is post stained with fluorescently labeled streptavidin and washed again. The array is then scanned to measure fluorescence signal at each spot on the array.