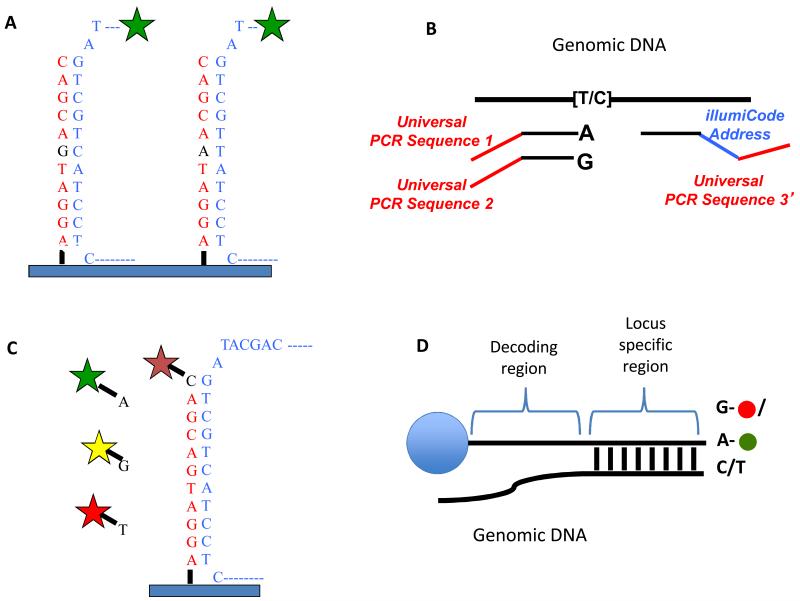
Figure 4.
SNP detections strategies for arrays. A) Allele discrimination by hybridization – Oligos that are complimentary to each allele are placed on the array and labeled genomic DNA is hybridized to the array. The variant position is placed in the center of the oligo (typically 25bp on Affymetrix arrays) as this position has the greatest affect on hybridization. Typically, multiple array positions are used for each allele to improve signal to noise. B) Illumina’s “Golden Gate Assay”- two allele specific oligos are each tailed with a different universal primer (1 and 2) and hybridized in solution to genomic DNA. A third oligo that is complementary to the same locus is tailed with a “barcode” sequence and a third universal primer (3). Polymerase is used to extend the allele specific primers across the genomic sequence and the extended products are ligated to the third oligo. PCR is performed using primers complimentary to universal sequences 1, 2 and 3. The PCR primers complimentary to the universal sequences 1 and 2 are labeled with a unique fluorophore. The barcode sequence on the third oligo allows the PCR product to be uniquely detected on an array containing oligos complimentary to the barcode sequence. The use of multiple barcodes (one for each locus of interest) allows the assay to be multiplexed to sample many loci. C) Arrayed primer extension (APEX) – In this assay, the array contains DNA oriented with the 5′ end attached to the array and the 3′ end stopping one nucleotide short of the SNP. Genomic DNA is fragmented and hybridized to the array and the oligo on the array is extended in single nucleotide dye terminator sequencing reaction. D) Illumina’s Infinium assay – This assay is similar to the APEX assay except that the oligo to be extended is on a bead and the single nucleotide that is added is labeled with a nucleotide specific hapten as opposed to a fluorophore. The haptens are then detected by staining with fluorescently labeled proteins that bind each hapten.