Figure 6.
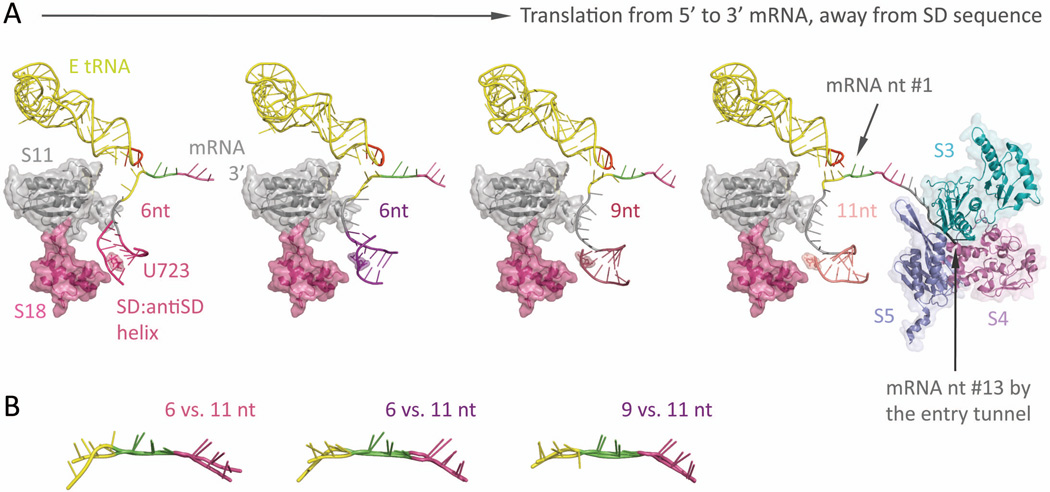
The springy Shine-Dalgarno•anti Shine-Dalgarno mini-helix (PDB: 2HGR,54 2QNH,52 6-nt; 3I8G,36 9-nt; 2HGP,54 11-nt) (A) Left-most: Around the mRNA 5’-end exit pore, a ~6-bp long SD•antiSD mini-helix (thin helical cartoon in magenta) resides within a pocket, surrounded by ribosomal proteins S11 and S18 (semitransparent space filling, with ribbon; in gray and magenta, respectively) from the 30S. Left two figures: The mini-helix (in magenta and purple) readily forms while the ribosome translates only 6-nt downstream from it (P-site codon colored in green, A-site codon in violet). Under such short spacing, the backbone of E-site codon (highlighted in yellow) is stretched and distinctively distorted from proper basepairing toward the E-tRNA anticodon (residue 34–36 highlighted in red, the rest in yellow); meanwhile, the center of the mini-helix roughly sits on top of the highly conserved residue, U723 (semitransparent spacefilling, with sticks; same color as the mini-helix), though the detailed orientation of the helix may vary slightly. Right two figures: As the ribosome translates forward (seen as the mini-helix sliding away from the residue U723), the nucleotide spacing increases (9-nt, middle right; 11-nt, right-most), thus relaxing the E-site codon (yellow) to resume hydrogen bonding with the anticodon (red). This illustrates how the formation of a SD•antiSD mini-helix, as well as its spacing between the codon•anticodon binding sites, can tune the availability of E-site condon•anticodon basepairing, which directly interferes with the mRNA registry and reading frame maintenance during translation. Right-most: The mRNA entry tunnel is composed of ribosomal proteins S3, S4, and S5 (semi-transparent space-filling, with ribbon; in cyan, dark pink, and slate, respectively). Counting from the first base in the Psite codon, nucleotide #13–15 would be located at the mRNA entry pore. This suggests a minimal spacing of 7 to 9-nt between a slippery sequence placed at the P- and A-site and a downstream hairpin at the entry pore. (B) Overlays of the decoded mRNA: various stretched forms of the E-site codon (at spacing of 6-nt and 9- nt) versus the relaxed form (11-nt).