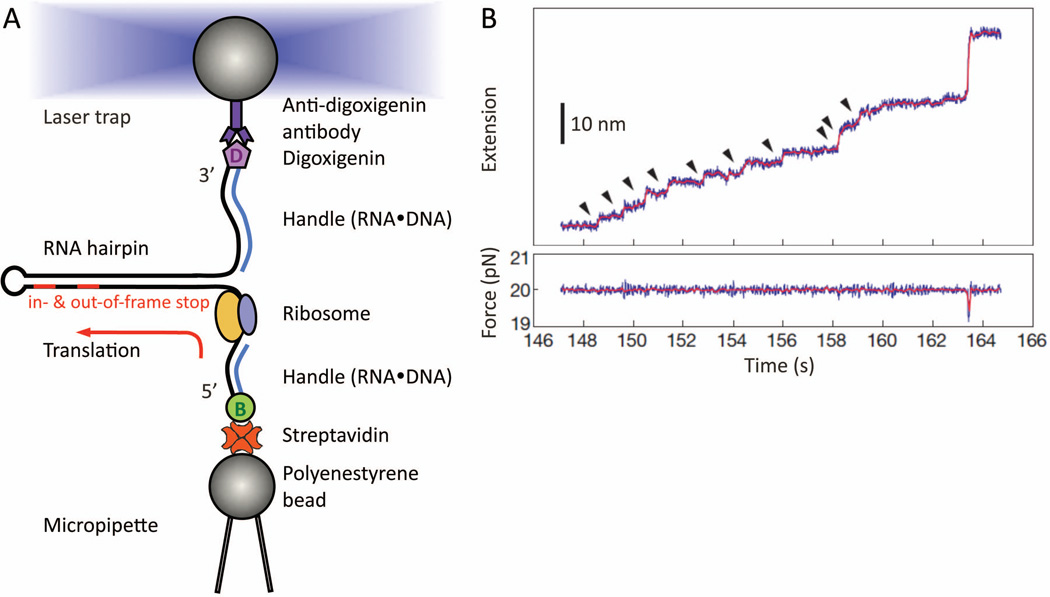
Figure 7.
Real-time step-by-step translation in an optical-tweezers experiment (A) Schematic drawing of the hairpin helicase assay used to follow single-ribosome translation. A selfcomplementary mRNA template (black line), which forms a hairpin, is held by its two ends. The tethering is done by attaching the RNA ends hybridized with either biotin- or digoxiginin- labeled DNA oligos, to either streptavidin- or anti-digoxigin antibody- coated polystyrene beads. While the bottom bead is fixed by suction onto the micropipette tip, the other is controlled by the focused laser trap. This allows us to adjust the force applied to the mRNA, thus tuning the mechanical stability of the hairpin that the ribosome will translate and unwind in 3-bp per codon-step. Note that no modifications on the ribosome, or other translation-essential components, are made. At the end of translation, the residual mRNA hairpin size provides a crosscheck confirming which codon the ribosome has reached, e.g. either the in- or out-of-frame stop codon on a frameshift-promoting mRNA. (B) Example of a step-pause-step translation trajectory from a single ribosome recorded as a function of time. This is captured as a series of fixed-size step-wise mRNA lengthening, i.e. extension, over time (top panel, ~2.7 nm per codon translated at ~20 pN). Here, a total of ten consecutive translation elongation cycles (arrow-highlighted) has been successfully followed in real-time. Each increment (vertical step) signifies the EF-G•GTP catalyzed ribosome translocation. If a frameshift occurs in parallel with the translocation step, a different extension step size will be observed. (Panel B is reproduced with permission from Figure 2A in reference 56.)