Figure 1. Genome-wide siRNA screen identifies determinants of resistance to erlotinib treatment.
A. Schematic diagram of the genome-wide siRNA screens performed in PC9 cells. Two sets of 384-well plates were reverse transfected with the genome-wide siRNA library and after 48hr treated with 30 nM erlotinib or left untreated (control) for 72 hours. Cells were fixed, stained with DAPI, and cell number was quantified on a microplate cytometer to obtain a Z-score for each siRNA in both conditions. Median Z-scores from the triplicate experiments are presented on the graph. siRNAs desensitizing cells to the drug are indicated in red (residual Z-score ≥ 2.0) and sensitizing siRNAs are indicated in blue (residual Z-score ≤ −2.0), whereas siRNAs that killed cells in the absence of the drug (control Z-score ≤ −2.0) are indicated in grey, and siRNAs that have no effect on modulating the response of cells to erlotinib (-2.0 < residual Z-score < 2.0, with control Z-score > 2.0) are indicated in black.
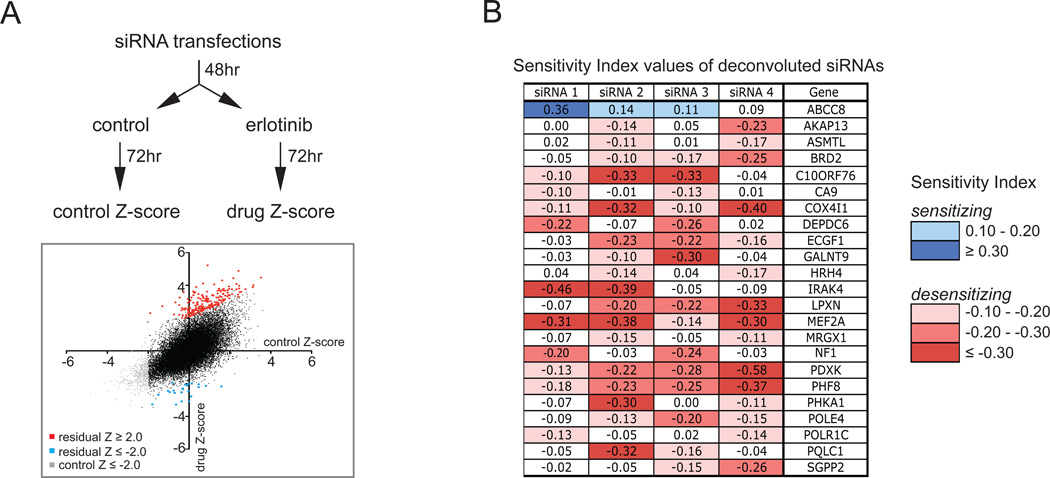
B. Tabulated sensitivity index-values of the deconvoluted siRNAs targeting 23 genes, of which two out of the four deconvoluted siRNAs had a substantial sensitizing (≥ 0.10) or desensitizing (≤ −0.10) effect on cell viability when cultured in the presence of erlotinib.