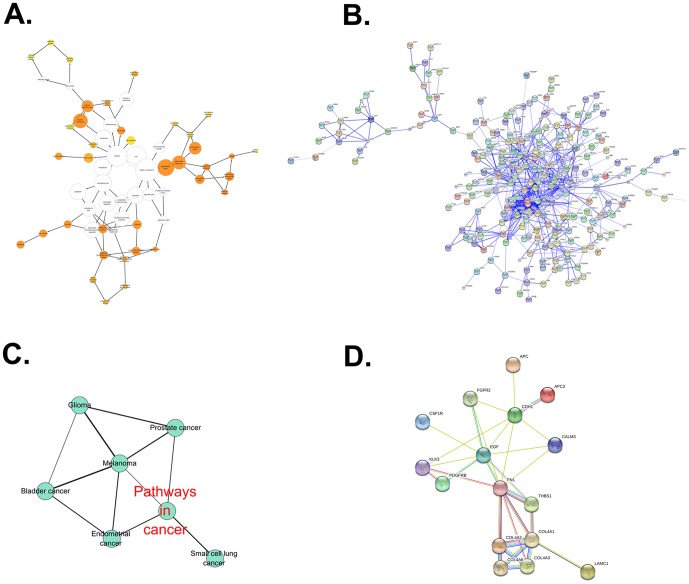
Figure 1. A cancer-associated PPI network in urine was constructed using a bioinformatics approach.
(A) A total of 592 high-abundance urinary proteins in our urinary proteome database were selected according to the criteria of having >4 unique peptides and a spectra number >10. Those proteins were analyzed using the BinGO plug-in in Cytoscape software, and 373 proteins were annotated as being “extracellular region” and “plasma membrane.” A map of the cellular component is shown. (B). The protein-protein interaction networks of the 373 proteins were constructed by STRING. A PPI network, which was composed of 312 nodes and 1779 interactions with removal of single nodes, was obtained. (C) The PPI network was analyzed by Cytoscape software with the ClueGO+Cluepedia plug-in. Six cancer-associated enriched KEGG terms were obtained and shown. (D) A cancer-associated urinary PPI network comprised of 15 urinary proteins that were associated with the 6 cancer-associated terms were constructed by STRING.