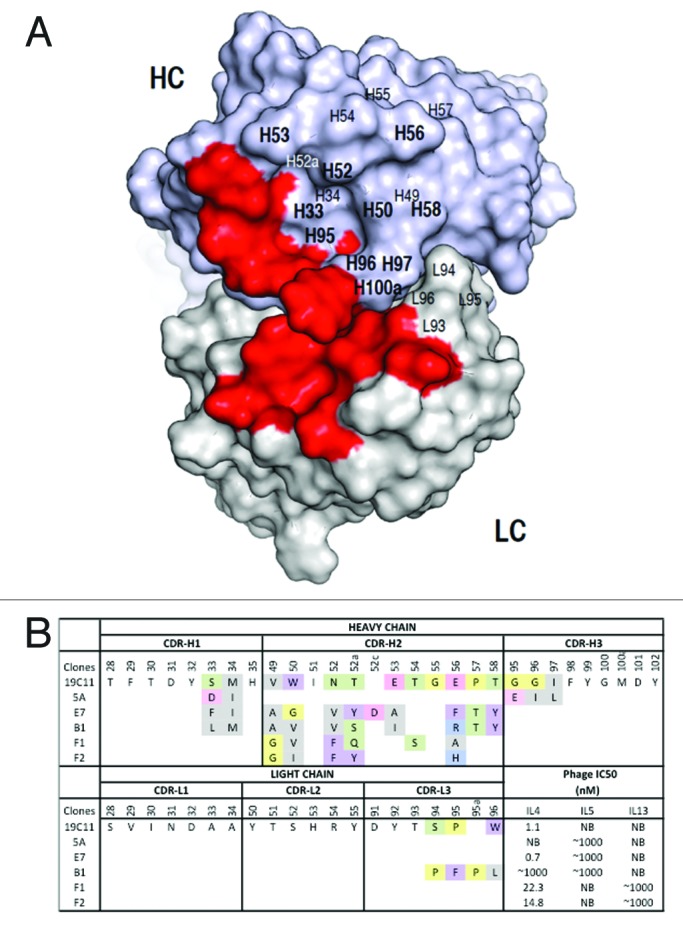
Figure 2. Dual specific clones isolated from phage libraries of h19C11 variants. (A) The structural model of hu19C11 was generated with MOE using the PDB entries 3SQO and 3BE1 as templates for the heavy and light chain, respectively. Residues important for IL4 binding, LC: 31, 32, 50, 53, 91, 92 and HC: 31, 32, 96, 98, 99 (in Kabat numbering), are colored in red on the model structure in a top down view of the antigen-binding site. Libraries of hu19C11 Fab variants displayed on phage were generated by site directed mutation. HC residues that were allowed all 20 amino acids with preference for wild type residues are labeled in bold while other HC and LC residues as labeled were allowed limited mutation to mimic natural diversity. (B) CDR sequences of selected clones including anti-IL5 specific (5A), anti-IL4/5 specific (E7, B1) and anti-IL4/13 specific (F1, F2) are shown with mutations from the parent hu19C11wt. To show the change of property upon mutation, sites included in the randomization scheme were colored according to their properties: Y, W, F in purple, hydrophobic L, I, V, A, M in gray, basic K, R, H in blue, acidic D, E in pink, polar S, T, N, Q in green, and P, G in yellow. The relative antigen binding affinity as measured by IC50 of phage competition assays are shown. Fab displaying phage was first incubated with serial dilutions of the respective antigens in solution for 2 h, unbound phage was then briefly captured by antigen coated ELISA wells and detected with anti-M13 HRP conjugate. The concentration of antigen inhibiting 50% of phage binding to antigen coated well is calculated as IC50. NB denotes no detectable binding signal of phage clone to the antigen coated wells.
