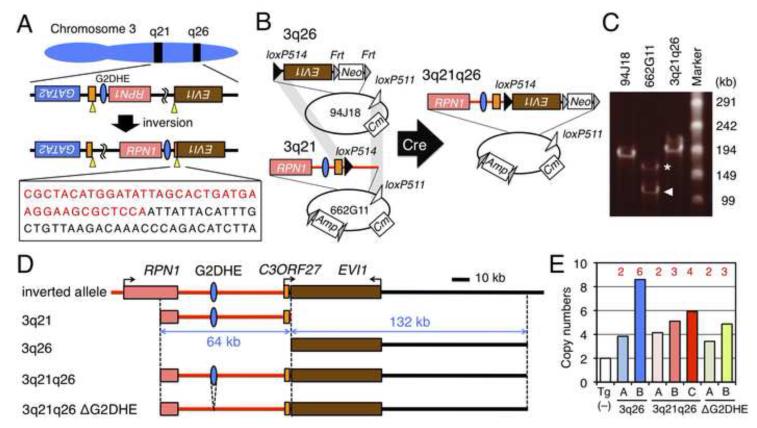
Figure 2. Generation of 3q21q26 mice by linking two BACs.
(A) Chromosomal inversion of the region between breakpoints (yellow arrowheads) in EVI1 (brown box) and C3ORF27 (orange box) genes in MOLM-1 cells. The boxed sequence shows the breakpoint between 3q21 (red) and 3q26 (black). (B) Strategy for linking two BAC clones. Human BACs RP11-94J18 (94J18) and RP11-662G11 (662G11) contain 3q26 and 3q21, respectively. We used site-specific Cre-mediated recombination to link the two BAC clones by simultaneous intermolecular homologous recombination between the loxP514 (black triangles) and loxP511 (white triangles) sites. Neo, Cm and Amp indicate the neomycin-, chloramphenicol- and ampicillin-resistant genes, respectively. (C) Pulse-field gel electrophoresis of BAC clones. We found two types of 662G11 clones; full-length (asterisk) and partially deleted (arrowhead) clones. (D) Schematic representations of the inverted allele, 3q21, 3q26, 3q21q26 and 3q21q26ΔG2DHE BAC transgenes. Red and black lines represent contributions from 3q21 and 3q26, respectively. (E) Copy numbers of integrated transgenes. Copy numbers were determined by q-PCR using Tg (−), 3q26 (lines A and B), 3q21q26 (lines A, B and C) and 3q21q26ΔG2DHE (lines A and B) mice (n=3-6). Red numbers depicted above the bar graphs represent copy numbers of transgenes.
See also Figure S2, Table S1 and Supplemental Experimental Procedures.