Figure 4.
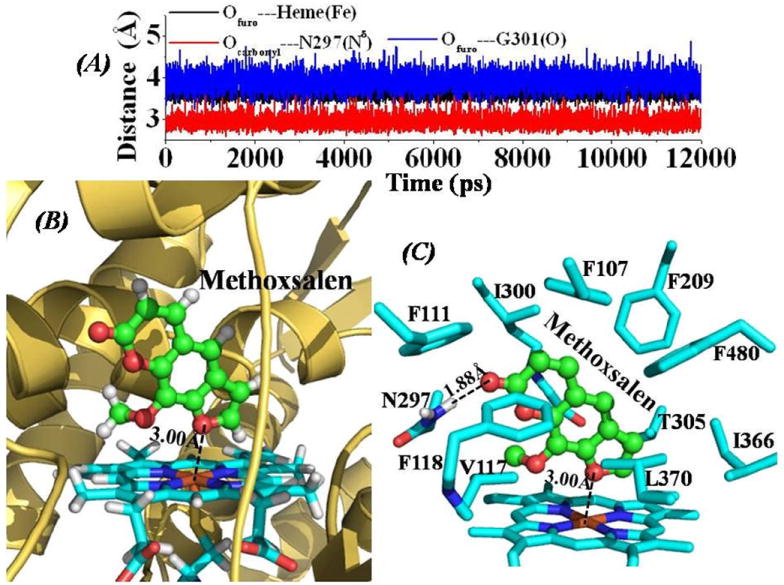
(A) Tracked distances along the MD trajectory for the CYP2A6-Methoxsalen binding structure. Ofuro---Heme(Fe) represents the distance between the oxygen atom on the furo group of Methoxsalen and the heme iron atom; Ocarbonyl---N297(Nδ) represents the distance between the carbonyl oxygen atom of Methoxsalen and the Nδ atom of the N297 side chain; Ofuro---G301(O) is the distance between the oxygen atom on the furo group of Methoxsalen and the carbonyl oxygen atom on the backbone of residue G310. (B) QM/MM-optimized CYP2A6-Methoxsalen binding structure (optimized at B3LYP/6-31G*:Amber8 level). The structure is represented as ribbon for CYP2A6, and stick style for the heme group, and Methoxsalen is displayed in ball-and-stick style. The dashed line represents the averaged distance between the furan oxygen atom and the heme iron atom based on the 10 QM/MM optimized structures. (C) Intermolecular interactions in the optimized CYP2A6-Methoxsalen binding structure. Residues from CYP2A6 within 5 Å of Methoxsalen are labeled and shown in stick style. The hydrogen-bonding interaction between the carbonyl oxygen atom of Methoxsalen and the Hδ atom of N297 side chain is represented as dashed line with labeled averaged distance based on the 10 QM/MM-optimized structures.
