A thylakoid membrane protein promotes the assembly of the protein complex that generates ATP in chloroplasts.
Abstract
The chloroplast F1Fo-ATP synthase/ATPase (cpATPase) couples ATP synthesis to the light-driven electrochemical proton gradient. The cpATPase is a multiprotein complex and consists of a membrane-spanning protein channel (comprising subunit types a, b, b′, and c) and a peripheral domain (subunits α, β, γ, δ, and ε). We report the characterization of the Arabidopsis (Arabidopsis thaliana) CONSERVED ONLY IN THE GREEN LINEAGE160 (AtCGL160) protein (AtCGL160), conserved in green algae and plants. AtCGL160 is an integral thylakoid protein, and its carboxyl-terminal portion is distantly related to prokaryotic ATP SYNTHASE PROTEIN1 (Atp1/UncI) proteins that are thought to function in ATP synthase assembly. Plants without AtCGL160 display an increase in xanthophyll cycle activity and energy-dependent nonphotochemical quenching. These photosynthetic perturbations can be attributed to a severe reduction in cpATPase levels that result in increased acidification of the thylakoid lumen. AtCGL160 is not an integral cpATPase component but is specifically required for the efficient incorporation of the c-subunit into the cpATPase. AtCGL160, as well as a chimeric protein containing the amino-terminal part of AtCGL160 and Synechocystis sp. PCC6803 Atp1, physically interact with the c-subunit. We conclude that AtCGL160 and Atp1 facilitate the assembly of the membranous part of the cpATPase in their hosts, but loss of their functions provokes a unique compensatory response in each organism.
The majority of cellular energy is stored in the form of ATP synthesized by the ubiquitous F1Fo-ATP synthase (F1 stands for coupling factor 1, Fo for coupling factor o), which is found in the energy-transducing membranes of bacteria, mitochondria, and chloroplasts. The chloroplast F1Fo-ATP synthase/ATPase (cpATPase) is a rotary motor that is responsible for coupling ATP synthesis (and hydrolysis) to the light-driven electrochemical proton gradient. The cpATPase comprises two physically separable parts, chloroplast coupling factor o (CFo), which is an integral membrane-spanning proton channel, and chloroplast coupling factor 1 (CF1), which is located peripheral to the membrane and contains the catalytic site(s) for reversible ATP synthesis (for review, see von Ballmoos et al., 2009). CFo comprises four different subunit types, designated b (synonymously, I or AtpF), b′ (II or AtpG), c (III or AtpH), and a (IV or AtpI), and contains one each of subunits a, b, and b′ and a ring made up of 14 copies of subunit c. CF1 comprises five different subunits, α (AtpA), β (AtpB), γ (AtpC), δ (AtpD), and ε (AtpE), and its subunit composition is α3β3γδε (for review, see von Ballmoos et al., 2009).
The passage of protons through the CFo motor drives rotation of the ring of c-subunits, which together form a rotor. The c-ring is connected to subunit γ, and rotation of γ causes conformational changes in the catalytic nucleotide-binding sites of the CF1 motor, resulting in the synthesis and release of ATP (for review, see Okuno et al., 2011). This process is made possible by the fact that CF1 and CFo are physically connected by two stalks, a central one containing the ε- and γ-subunits and a peripheral one made up of δ, b, and b′ (for review, see Böttcher and Gräber, 2000; Weber, 2007). There are six nucleotide-binding sites in CF1, one at each of the αβ-subunit interfaces about halfway along the vertical axis of the hexamer. Three of the sites are located primarily on the β-subunits and are catalytic; the other three are noncatalytic and probably regulatory. While the three-dimensional structure of the α3β3 hexamer in chloroplasts has been solved to a resolution of 3.2 Å (Groth and Pohl, 2001), the structure of the entire CFo has not yet been determined. However, the conformation of the ring-forming part of CFo from spinach (Spinacia oleracea) chloroplasts has been defined and found to consist of 14 c-units (Vollmar et al., 2009), whereas the c-ring of the ATP synthase from the cyanobacterium Spirulina platensis contains 15 units (Pogoryelov et al., 2009).
Similar to other thylakoid multiprotein complexes like PSII and PSI as well as the cytochrome b6f complex (Cyt b6f), the assembly of the ATP synthase must be tightly regulated. Moreover, the variable stoichiometry of the constituents of F1 (three α/β-subunits versus one each of γ, δ, and ε) and Fo (10–15 c-subunits versus one each of a, b, and b′) requires coordination of the expression of the corresponding genes. This is particularly important in eukaryotes, where the genes are located in different compartments, for instance, in the case of the cpATPase, in the plastid (for α, β, ε, a, b, and c) and the nucleus (for b′, γ, and δ).
The assembly of ATP synthase has been most extensively studied in Saccharomyces cerevisiae mitochondria, leading to the identification of several factors involved in this process (for review, see Rak et al., 2009). Thus, three proteins in yeast are known to be involved in the assembly of the α3β3 hexamer of F1. Atp11p (Ackerman and Tzagoloff, 1990a; Wang and Ackerman, 1996) and Atp12p (Ackerman and Tzagoloff, 1990a; Wang and Ackerman, 1998) code for mitochondrial proteins that interact with the β- and α-subunits, respectively, to promote their assembly into the oligomeric F1-ATPase, and the absence of either protein causes the α- and β-subunits to aggregate into insoluble inclusion bodies in the mitochondrial matrix. Lack of the third protein, FORMATION OF MITOCHONDRIAL COMPLEXES1 (Fmc1p), is associated with aggregation of the α- and β-subunits under heat stress, suggesting that Fmc1p is required for correct folding of Atp12p at elevated temperatures (Lefebvre-Legendre et al., 2001). Originally, the c-ring was assumed to form spontaneously (Arechaga et al., 2002), but subsequent studies have indicated that the assembly of this structural component is also a protein-assisted process. Thus, Atp25p is required for both the synthesis of the c-subunit and its oligomerization into a ring structure of the proper size (Zeng et al., 2008). Moreover, Atp10p (Ackerman and Tzagoloff, 1990b), Atp23p (Osman et al., 2007), and OXIDASE ASSEMBLY1 (Oxa1p) (Jia et al., 2007) are involved in Fo assembly in yeast mitochondria.
In prokaryotes, two ATP synthase assembly factors have been described in detail. The membrane protein insertase YidC belongs to the Oxa1 family, is required in vitro for the membrane insertion of subunit c, and assists in the formation of the c-ring from monomers (van der Laan et al., 2004; Kol et al., 2008). In bacterial genomes, the atp1/uncI genes typically precede the genes encoding the structural subunits of the F1Fo-ATP synthase (for review, see Kol et al., 2008). Moreover, in Synechocystis sp. PCC6803, sll1321/atp1 is coordinately expressed with the seven other genes in the ATP synthase operon (Grossman et al., 2010), implying that Sll1321/Atp1 might have a function associated with the ATP synthase. The genes atp1 and uncI code for small proteins; for instance, Synechocystis sp. PCC6803 Sll1321 has 117 amino acids, and Escherichia coli UncI has 130 amino acids. The function of Atp1/UncI has long remained elusive because deletion of uncI in E. coli results merely in a slightly reduced growth yield (Gay, 1984), indicating that the protein is not essential for the formation of the F1Fo-ATP synthase complex. Similarly, in the alkaliphilic Bacillus pseudofirmus OF4, Atp1/UncI is not absolutely required for ATP synthase function, and a B. pseudofirmus strain deleted for the atp1 gene could still grow nonfermentatively and its purified ATP synthase had a c-ring of normal size (Liu et al., 2013). Recently, a hybrid F1Fo (F1 from Bacillus PS3 and Fo from Propionigenium modestum) was expressed in E. coli. In this system, P. modestum Atp1/UncI was found to be indispensable for c-ring formation and coupled ATPase activity (Suzuki et al., 2007). Similarly, functional production of the Na+ F1Fo-ATP synthase from Acetobacterium woodii in E. coli required the A. woodii atp1/uncI gene for proper assembly (Brandt et al., 2013). Moreover, because subunit c monomers, as well as assembled c-rings, can be copurified together with P. modestum UncI/Atp1 (Suzuki et al., 2007) and the oligomerization of P. modestum c-subunits into c11-rings is mediated by Atp1/UncI in vitro (Ozaki et al., 2008), Atp1/UncI seems to play a role in c-ring assembly for some bacterial ATP synthases.
In plants and green algae, regulation of the biogenesis of the cpATPase is well understood at the level of translation of CF1 subunits (Drapier et al., 2007). Thus, synthesis of the nucleus-encoded subunit γ is required for sustained translation of the chloroplast-encoded subunit β, which in turn transactivates the translation of chloroplast-encoded subunit α. Translational down-regulation of subunit β or α, when not assembled, involves the 5′ untranslated regions (UTRs) of their own mRNAs, pointing to control at the level of translation initiation. In addition, a negative feedback exerted by α/β assembly intermediates on the translation of subunit β can be released when subunit γ assembles with α3β3 hexamers.
Our knowledge of the nature of true assembly factors for the cpATPase is scarce. So far, only the ALBINO3 homolog Alb4 protein, which can functionally substitute for YidC in E. coli, has been shown to play a role in the biogenesis of the cpATPase, possibly by stabilizing or promoting the assembly of CF1 during its attachment to the CFo portion (Benz et al., 2009). Thus, Alb4-Oxa1p-YidC represents an ATP synthase assembly factor family that is conserved between prokaryotes, yeast, and plants. For the bacterial Atp1/UncI protein, one homolog exists in yeast, Vma21p, which is an integral membrane protein localized to the endoplasmic reticulum and is required for vacuolar H+-ATPase biogenesis (Graham et al., 1998).
In this study, we have identified and characterized a knockout mutant for Arabidopsis (Arabidopsis thaliana) CGL160, a protein that displays moderate similarity to prokaryotic Atp1/UncI proteins in its C-terminal domain. AtCGL160 is required for the efficient assembly of the cpATPase, but lack of AtCGL160 in Arabidopsis has more severe effects on cpATPase assembly than those reported in the literature for inactivation of its prokaryotic relatives and can be located to the assembly of c-subunits into the membranous subcomplex. AtCGL160 physically interacts with the c-subunit of CFo, and, interestingly, Atp1 can replace the C-terminal part of AtCGL160 in such interactions, indicating that the function of Atp1 and CGL160 proteins is conserved.
RESULTS
The GreenCut Protein AtCGL160 Contains a Short Atp1/UncI-Like Domain
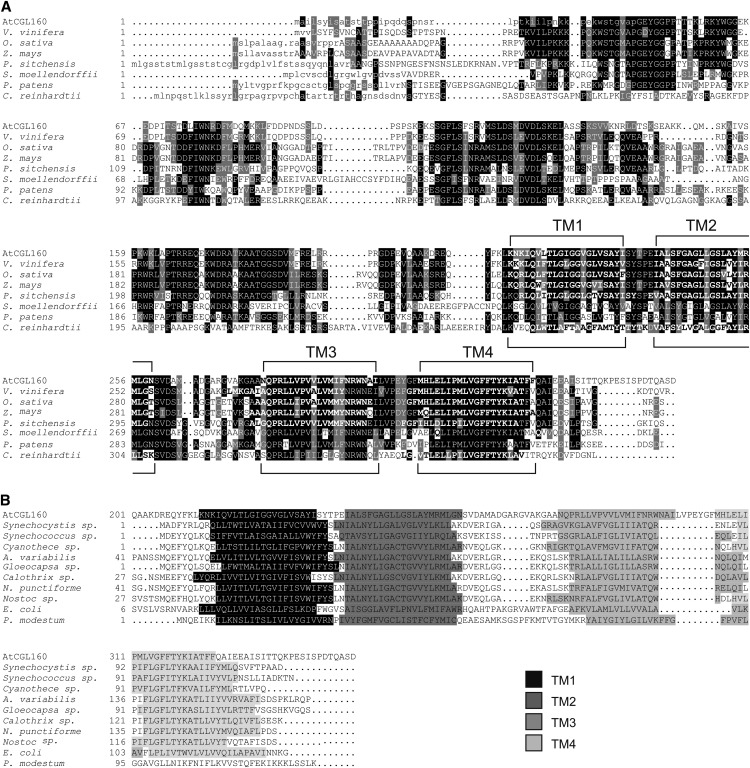
In the course of a systematic screen for novel photosynthesis-relevant proteins in Arabidopsis, a set of genes was considered that (1) are shared by photosynthetic eukaryotes from the green lineage but are not found in nonphotosynthetic eukaryotes (the so-called “GreenCut” proteins; Merchant et al., 2007; Grossman et al., 2010) and (2) show a photosynthesis-specific mRNA expression profile (Biehl et al., 2005; DalCorso et al., 2008; Takabayashi et al., 2009; for coregulated photosynthesis-related genes, see Supplemental Table S1). One of these genes is AT2G31040, the Arabidopsis equivalent of the Chlamydomonas reinhardtii gene CGLD22 (Merchant et al., 2007), which was later renamed CGL160 (Karpowicz et al., 2011). In the following, we designate AT2G31040 as AtCGL160 and its C. reinhardtii ortholog as CrCGL160. AtCGL160 codes for a protein of 350 amino acids containing an N-terminal chloroplast transit peptide (cTP) of 46 amino acids, as predicted by ChloroP (Emanuelsson et al., 1999), such that the mature protein should have a molecular mass of approximately 34 kD. AtCGL160, like its counterparts from other vascular plants, mosses, and green algae, contains four transmembrane (TM)-spanning α-helices (hereafter named the “membrane domain”) in its C-terminal region (Fig. 1A). CGL160 from the moss Physcomitrella patens shows 53%/40% similarity/identity to AtCGL160, while the values for CrCGL160 are lower (38%/25% similarity/identity). Cross-species sequence conservation (Fig. 1A) is more pronounced in the membrane domain (84%/25.7% consensus positions/identity positions; amino acids 209–350 of AtCGL160) than in the N-terminal segment (69.9%/6.7%; amino acids 47–208 of AtCGL160). Here, “identity positions” refers to positions with identical amino acids in each species, whereas “consensus positions” refers to positions at which at least 50% of species examined show identical or similar residues.
Figure 1.
Sequence alignment of AtCGL160 and its homologs from other species. A, The sequence of the AtCGL160 protein was compared with related sequences from the eukaryotes grape (Vitis vinifera), rice (Oryza sativa), maize (Zea mays), Picea sitchensis, Selaginella moellendorffii, Physcomitrella patens, and C. reinhardtii. Predicted cTPs are indicated by lowercase letters and TM-spanning α-helices by boldface letters. Sequence similarity and identity in at least 50% of the sequences are highlighted by gray and black shading, respectively. The positions of the four TM regions (TM1–TM4) are indicated. B, The sequence of the C-terminal membrane domain of AtCGL160 (AtCGL160201-350) was compared with related prokaryotic sequences. AtCGL160201-350 was aligned with Atp1/UncI sequences from the cyanobacteria Synechocystis sp. PCC6803, Synechococcus sp., Cyanothece sp., Anabaena variabilis, Gloeocapsa sp., Calothrix sp., Nostoc punctiforme, and Nostoc sp. and with sequences from P. modestum and E. coli. The four TM regions are highlighted in different shades of gray. Note that for UncI from P. modestum, only three TM regions are predicted.
The GreenCut includes genes found in photosynthetic eukaryotes but not in nonphotosynthetic organisms, whether prokaryotes or eukaryotes. Nevertheless, the conserved C-terminal portion of AtCGL160 is distantly related to the products of bacterial atp1/uncI genes, such as the atp1/sll1321 gene in Synechocystis sp. PCC6803 (Grossman et al., 2010; Karpowicz et al., 2011) and the uncI gene in E. coli (Walker et al., 1984). Although sequence alignments of the C-terminal membrane domain of AtCGL160 with the products of atp1/uncI genes from different cyanobacteria and E. coli showed only moderate overall similarity, the relative positions of four predicted TM-spanning α-helices coincided (Fig. 1B). This suggests that the secondary structure of this domain is conserved between eukaryotic CGL160 and bacterial Atp1/UncI proteins. UncI from P. modestum contains only three predicted TM-spanning α-helices, but its membrane domain still shows some sequence conservation (similarity/identity, 33.6%/18.6%) with that of AtCGL160 (Fig. 1B).
AtCGL160 Is Required for Efficient Photosynthesis
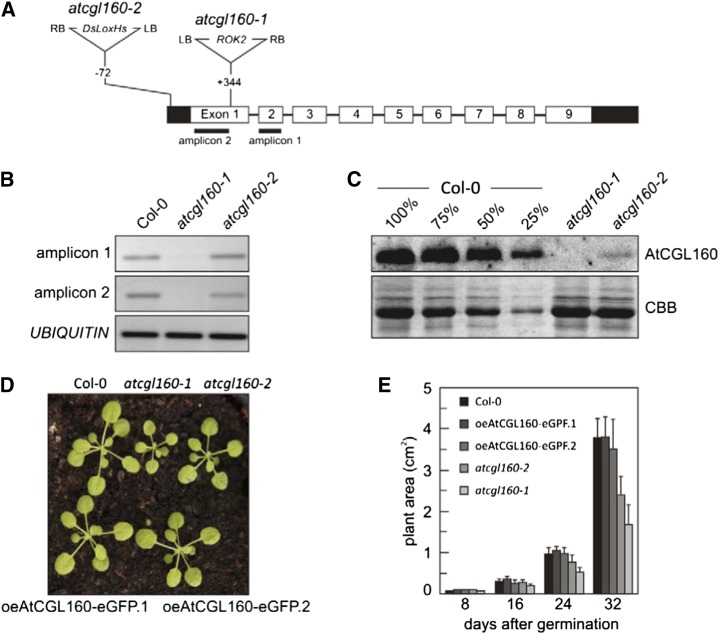
Two different Arabidopsis transfer DNA (T-DNA) insertion lines for AtCGL160 were identified in the SIGnAL database (http://signal.salk.edu/cgi-bin/tdnaexpress) and obtained from the Nottingham Arabidopsis Stock Centre. The sites of the T-DNA insertions were determined by PCR amplification and sequencing of the regions flanking the T-DNA borders. In atcgl160-1 (SALK_057229), the ROK2 T-DNA was inserted in the first of the nine exons of AtCGL160, whereas the DsLoxHs T-DNA in atcgl160-2 (WiscDsLoxHs024_02B) was inserted in the 5′ UTR (Fig. 2A). Reverse transcriptase-PCR analyses were performed to examine the effects of the T-DNA insertions on AtCGL160 transcript abundance in the mutant lines. Neither complementary DNA (cDNA) from an amplicon located 3′ of the insertion (amplicon 1 in Fig. 2A) nor from an amplicon located 5′ of the insertion (amplicon 2) could be obtained from atcgl160-1 plants, whereas both portions of AtCGL160 transcripts were detectable in atcgl160-2 (Fig. 2B). To analyze AtCGL160 expression at the protein level, the N-terminal portion of AtCGL160 was heterologously expressed in, and purified from, E. coli, and an antibody raised against it was employed in immunoblot assays of thylakoid preparations from wild-type (Columbia-0 [Col-0]) and mutant (atcgl160-1 and atcgl160-2) plants (Fig. 2C). As expected, the AtCGL160 protein failed to accumulate in atcgl160-1 thylakoid membrane samples, but protein amounts corresponding to less than 25% of wild-type AtCGL160 levels were detected in atcgl160-2. The sizes of atcgl160-1 and, to a lesser extent, atcgl160-2 plants were clearly reduced relative to wild-type plants (Fig. 2D). Accordingly, the growth rate of atcgl160-1, as determined by leaf area measurements (Leister et al., 1999), was markedly reduced, whereas an effect of the atcgl160-2 mutation on plant growth only became evident in older plants (32 d after germination; Fig. 2E).
Figure 2.
Characterization of the T-DNA mutant lines atcgl160-1 and atcgl160-2. A, Structure of the AtCGL160 gene in Arabidopsis and positions of the T-DNA insertions in the mutant lines. Left and right T-DNA borders are indicated by LB and RB, respectively. Exons are numbered (1–9) and shown as white rectangles, and UTRs are shown as black rectangles. The locations of the amplicons used in B are indicated below the corresponding exons. Note that T-DNAs are not drawn to scale. B, AtCGL160 transcript abundance was analyzed by reverse transcriptase-PCR (25 cycles) of amplicons 1 and 2, the positions of which are shown in A. Primer annealing sites were selected to allow the amplification of cDNA fragments at locations 5′ and 3′ of the T-DNA integration site in atcgl160-1. C, Immunodetection of the AtCGL160 protein in wild-type (Col-0) and mutant (atcgl160-1 and atcgl160-2) plants. Thylakoid membrane fractions (10 µg as determined by amido black staining) isolated from wild-type (Col-0) and mutant leaves, as well as diluted wild-type samples (75%, 50%, and 25%), were fractionated on SDS-PAGE gels and analyzed by immunoblotting employing an AtCGL160-specific antibody. As a loading control, Coomassie Brilliant Blue G-250 staining of transferred proteins on a PVDF membrane is shown (CBB). D, Growth phenotypes of wild-type (Col-0) and mutant (atcgl160-1 and atcgl160-2) lines as well as of two independently generated lines that overexpress AtCGL160-eGFP in the atcgl160-1 background (oeAtCGL160-eGFP.1 and oeAtCGL160-eGFP.2). E, Quantification of mean leaf area in wild-type (Col-0), mutant (atcgl160-1 and atcgl160-2), and overexpressor (oeAtCGL160-eGFP.1 and oeAtCGL160-eGFP.2) plants. Plants were grown under short-day conditions, and leaf area was measured at four time points (8, 16, 24, and 32 d after germination). Values are averages of 20 plants, and error bars show sd.
To confirm that the growth phenotype observed in atcgl160-1 results from knockout of the AtCGL160 gene, the wild-type AtCGL160 gene was fused to the enhanced green fluorescent protein (eGFP)-encoding reporter gene, placed under the control of the cauliflower mosaic virus 35S promoter, and introduced into atcgl160-1 plants. Two independently generated transgenic lines, designated oeAtCGL160-eGFP.1 and oeAtCGL160-eGFP.2, were further analyzed and found to overexpress AtCGL160-eGFP (Supplemental Fig. S1, A and B) and to display wild-type-like growth (Fig. 2E), implying that the atcgl160-1 growth phenotype is indeed caused by knockout of the AtCGL160 gene. Hence, all subsequent biochemical and physiological experiments were performed with the atcgl160-1 allele.
To examine the effect of AtCGL160 disruption on photosynthesis, the composition of the leaf pigments and fluorescence parameters for chlorophyll (Chl) a were analyzed. Pigments were extracted from light-adapted wild-type (Col-0), atcgl160-1, and oeAtCGL160-eGFP.1 leaves and analyzed by reverse-phase HPLC (Table I). The AtCGL160-eGFP overexpressor behaved like the wild type with respect to its leaf pigment content. With respect to total Chl (Chl a + b), the carotenoids lutein and carotene, as well as neoxanthin, the atcgl160-1 mutant also behaved similarly to the wild type. However, the size of the pool of violaxanthin + antheraxanthin + zeaxanthin was clearly increased in atcgl160-1 plants. Moreover, the levels of the deepoxidation products of violaxanthin, namely antheraxanthin and zeaxanthin, were dramatically increased in atcgl160-1, partly at the expense of violaxanthin, implying that atcgl160-1 plants suffer from light stress. The analysis of Chl a fluorescence showed that the maximum quantum yield of PSII in atcgl160-1 leaves was only slightly reduced relative to the wild type (Col-0; Table II), indicating that PSII is not impaired in atcgl160-1 thylakoids. However, the effective quantum yield of PSII, as well as the maximum electron transport rate (ETR) through PSII, was depressed in atcgl160-1 plants compared with wild-type plants (Fig. 3A; Table II). Nonphotochemical quenching of chlorophyll a fluorescence (NPQ) was increased in atcgl160-1, indicating that a larger fraction of the absorbed light energy is dissipated as heat (Fig. 3B). In addition, the reduction state of the plastoquinone pool was found to be higher in atcgl160-1 than in wild-type plants (Table II). The AtCGL160-eGFP overexpressor behaved essentially like the wild type with respect to its Chl a fluorescence parameters.
Table I. Pigment analyses of wild-type (Col-0), atcgl160-1, and oeAtCGL160-eGFP leaves.
Samples were harvested 6 h after the beginning of the light phase (80–100 µmol photons m−2 s−1) from 6-week-old plants grown in a climate chamber under short-day conditions. Pigments were extracted and quantified as described in “Materials and Methods.” Average values (n = 5) and sd were calculated, and values are given in pmol mg−1 fresh weight.
| Pigment | Col-0 | atcgl160-1 | oeAtCGL160-eGFP.1 | oeAtCGL160-eGFP.2 |
|---|---|---|---|---|
| Neoxanthin | 57 ± 2 | 58 ± 3 | 57 ± 9 | 55 ± 5 |
| Violaxanthin | 38 ± 2 | 21 ± 1 | 39 ± 6 | 36 ± 5 |
| Antheraxanthin | 1 ± 0 | 22 ± 1 | 1 ± 0 | 2 ± 1 |
| Zeaxanthin | 0 ± 0 | 25 ± 2 | 0 ± 0 | 1 ± 2 |
| Violaxanthin + antheraxanthin + zeaxanthin | 39 ± 2 | 68 ± 2 | 40 ± 6 | 39 ± 4 |
| Lutein | 178 ± 6 | 192 ± 13 | 184 ± 28 | 183 ± 22 |
| β-Carotene | 164 ± 5 | 157 ± 8 | 165 ± 27 | 162 ± 17 |
| Chl a | 1,400 ± 40 | 1,367 ± 55 | 1,387 ± 216 | 1,335 ± 120 |
| Chl b | 455 ± 15 | 467 ± 24 | 448 ± 70 | 439 ± 43 |
| Chl a + b | 1,855 ± 54 | 1,834 ± 77 | 1,835 ± 286 | 1,775 ± 163 |
| Chl a/b ratio | 3.07 ± 0.03 | 2.93 ± 0.08 | 3.09 ± 0.02 | 3.04 ± 0.03 |
Table II. Chl a fluorescence parameters of wild-type (Col-0), atcgl160-1, and oeAtCGL160-eGFP plants.
Leaves were exposed to 100 µmol photons m−2 s−1 for 15 min. The dark relaxation phase lasted 10 min. Averages values (n = 5–6) ± sd are provided. Fv/Fm, Maximal quantum yield of PSII; ΦII, effective quantum yield of PSII at 100 µmol photons m−2 s−1; 1-qP, excitation pressure.
| Parameter | Wild Type | atcgl160-1 | oeAtCGL160-eGFP.1 | oeAtCGL160-eGFP.2 |
|---|---|---|---|---|
| Fv/Fm | 0.81 ± 0.01 | 0.79 ± 0.01 | 0.81 ± 0.00 | 0.81 ± 0.00 |
| ΦII | 0.51 ± 0.02 | 0.41 ± 0.02 | 0.56 ± 0.03 | 0.55 ± 0.03 |
| 1-qP | 0.28 ± 0.03 | 0.36 ± 0.02 | 0.26 ± 0.02 | 0.28 ± 0.02 |
| NPQ | 0.59 ± 0.08 | 1.10 ± 0.06 | 0.41 ± 0.12 | 0.43 ± 0.12 |
| qE | 0.42 ± 0.07 | 1.00 ± 0.06 | 0.27 ± 0.13 | 0.27 ± 0.03 |
| qI | 0.21 ± 0.03 | 0.17 ± 0.02 | 0.19 ± 0.04 | 0.19 ± 0.03 |
Figure 3.
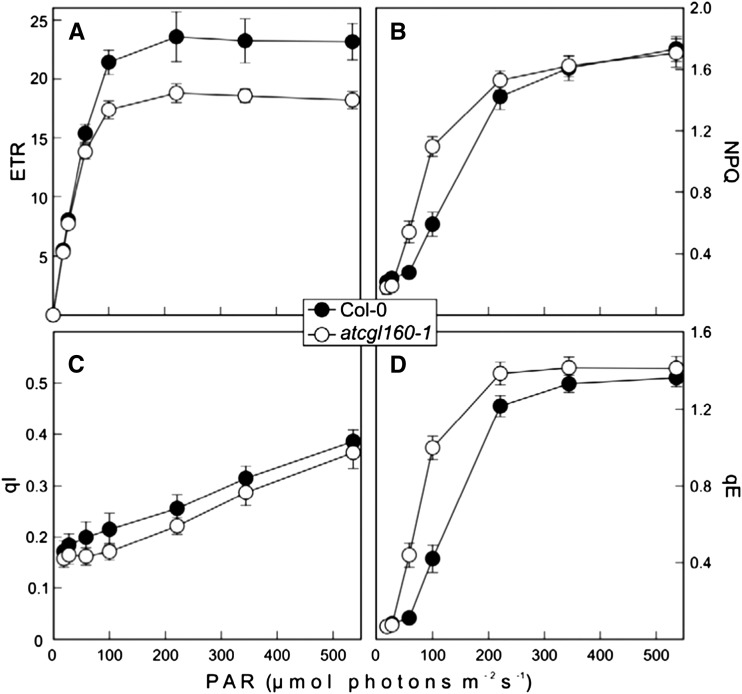
Effects of the atcgl160-1 mutation on photosynthetic electron flow and excess energy dissipation. ETR (in µmol electrons m−2 s−1; A), NPQ (B), qI (C), and qE (D) were determined in wild-type (Col-0) and atcgl160-1 leaves at seven different light intensities. Before the measurements, plants were dark incubated for 20 min. Maximal quantum yields were determined shortly before the onset of the light treatment. Leaves were exposed for 15 min to different light intensities (13, 22, 53, 95, 216, 339, and 531 µmol photons m−2 s−1) followed by a dark relaxation period of 10 min. ETR and NPQ values were calculated on the basis of fluorescence parameters measured during a saturating pulse at the end of each light interval, whereas qE and qI were determined on the basis of fluorescence parameters measured during a saturating pulse at the end of each dark relaxation period. Values are averages of six measurements, and error bars indicate sd.
In summary, lack of AtCGL160 is associated with the accumulation of antheraxanthin and zeaxanthin and higher levels of controlled heat dissipation under moderate light intensities. PSII appears to be unaffected by the inactivation of AtCGL160, whereas subsequent electron transfer steps are perturbed, causing overreduction of the plastoquinone pool.
Lack of AtCGL160 Increases Energy-Dependent Quenching of Chl Fluorescence
To analyze the contribution of the energy-dependent quenching of chlorophyll fluorescence (qE; or ΔpH-dependent quenching) and photoinhibitory quenching (qI) components of NPQ, dark-relaxation analyses of Chl fluorescence yields were performed at seven different light intensities (Fig. 3, C and D). The mechanisms responsible for the slow relaxation (more than 10 min) of Chl fluorescence yields are mainly attributed to the effects on qI, whereas fast relaxation (less than 10 min) reflects qE (Thiele et al., 1997). Photoprotective mechanisms arising from lumen acidification (protonation of PSII subunit PsbS and conversion of violaxanthin to zeaxanthin by the violaxanthin deepoxidase) are quantifiable by qE measurements. Therefore, an elevated qE value normally indicates an increase in lumenal proton concentrations.
Values of qI in atcgl160-1 and wild-type (Col-0) plants rose almost linearly with increasing light intensity; however, qI values for atcgl160-1 were always slightly lower than in the wild type (Fig. 3C). The qE values for atcgl160-1 and wild-type plants (Col-0) differed even under low light, but the greatest difference was observed at 100 µmol photons m−2 s−1, where qE in atcgl160-1 (at 1 ± 0.06) was more than twice as high as in the wild type (0.42 ± 0.07). Moreover, qE saturation was observed at lower light intensities (220 µmol photons m−2 s−1) for atcgl160-1 than for the wild type (340–540 µmol photons m−2 s−1; Fig. 3D).
To test whether wild-type-like photosynthesis was restored in the two AtCGL160-eGFP overexpressor lines, the quantum yield of regulated heat dissipation in PSII, Y(NPQ) (Kramer et al., 2004), was measured using an Imaging PAM system (Walz; Supplemental Fig. S1C). Both overexpressor lines exhibited wild-type-like Y(NPQ) values, corroborating the previous inference that the AtCGL160-eGFP fusion protein is functional in the two independently derived complementation lines.
Altogether, atcgl160-1 is characterized by a high-qE phenotype, which becomes manifest even under low light. This is also compatible with the rise in xanthophyll cycle activity indicated by the dramatically increased zeaxanthin and antheraxanthin levels in atcgl160-1.
AtCGL160 Is Required for the Normal Accumulation of cpATPase
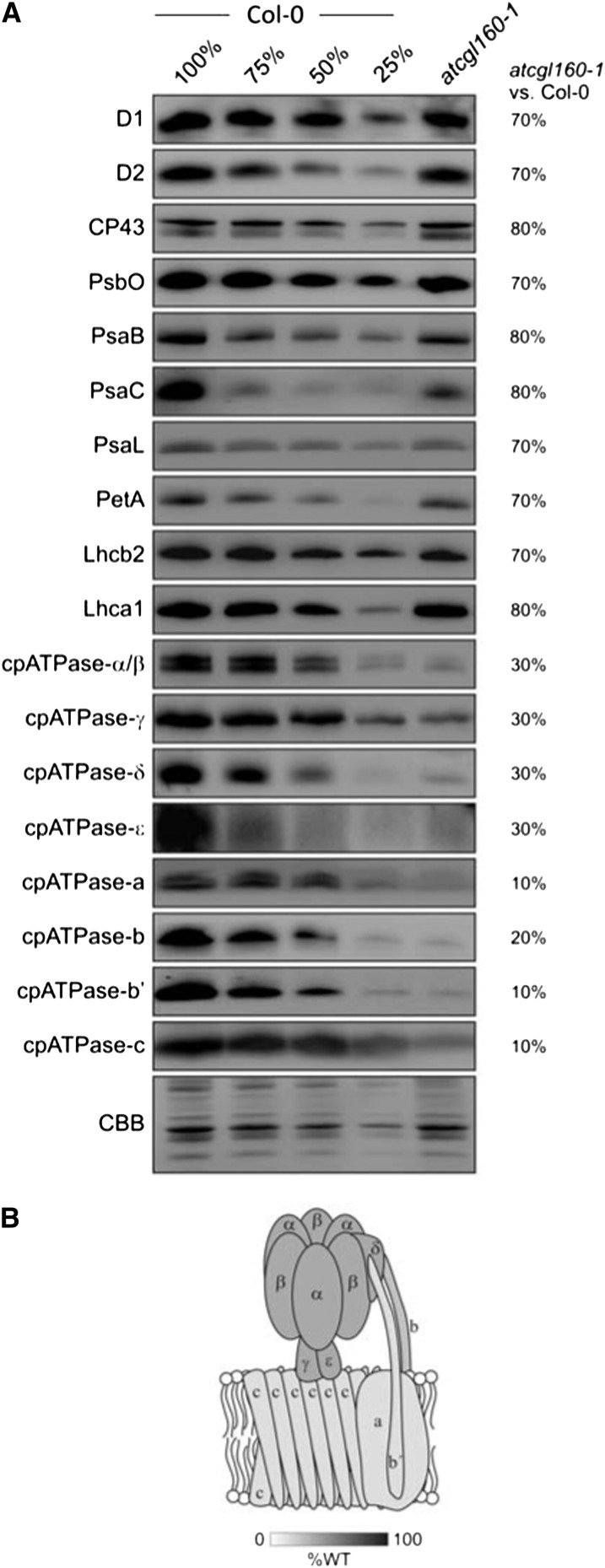
To analyze the influence of AtCGL160 on the abundance of the major thylakoid multiprotein complexes, the accumulation of marker proteins for the respective complexes was monitored (Fig. 4). In atcgl160-1 plants, PSII (represented by D1, D2, CP43, and PsbO), PSI (PsaB, PsaC, and PsaL), Cyt b6f (PetA), light-harvesting complex II (LHCII; represented by the PSII light-harvesting complex protein2 [Lhcb2]), and LHCI (Lhca1) were found to be reduced to 70% to 80%, 70% to 80%, 70%, 70%, and 80%, respectively of wild-type (Col-0) amounts. The most notable effect, however, concerned the cpATPase complex. Only 30% of wild-type amounts of CF1 subunits (α/β, δ, γ, and ε) and still lower levels (10%–20% of wild-type amounts) of CFo subunits (a, b, b′, and c) were detected in atcgl160-1 thylakoid membranes.
Figure 4.
AtCGL160 is required for the normal accumulation of cpATPase. A, Thylakoid membrane-enriched fractions were isolated from wild-type (Col-0) and atcgl160-1 plants, and aliquots corresponding to 10 μg of total protein were loaded in lanes Col-0 and atcgl160-1, together with serial dilutions of wild-type samples (75%, 50%, and 25%), and fractionated on SDS-PAGE gels. Antibodies specific for subunits of PSII (D1, D2, CP43, PsbO), PSI (PsaB, PsaC, PsaL), Cyt b6f (PetA), LHCs (Lhcb2, Lhca1), and the cpATPase (cpATPase-α/β, cpATPase-γ, cpATPase-δ, cpATPase-ε, cpATPase-a, cpATPase-b, cpATPase-b′, and cpATPase-c) were then used to identify proteins after transfer to PVDF membranes, and signals from atcgl160-1 samples were quantified relative to the wild type (relative values in percentage of wild-type values are given on the right). CBB, Coomassie Brilliant Blue G-250. B, The extent of depletion of the different subunits of the cpATPase in atcgl160-1 thylakoids (relative to the wild type [%WT]) is indicated by the shading of the schematic model of the complex.
The strong reduction in cpATPase content is in agreement with the high-qE phenotype of atcgl160-1, because both proton gradient-generating complexes (PSII and Cyt b6f) are much less affected than the cpATPase. Consequently, protons should accumulate in the lumen and trigger energy-dependent quenching mechanisms (Table I). Accordingly, reductions in the amounts of PSII, PSI, LHCs, and associated pigments can be interpreted as secondary effects of the relative lack of cpATPase.
AtCGL160 Is an Integral Thylakoid Protein
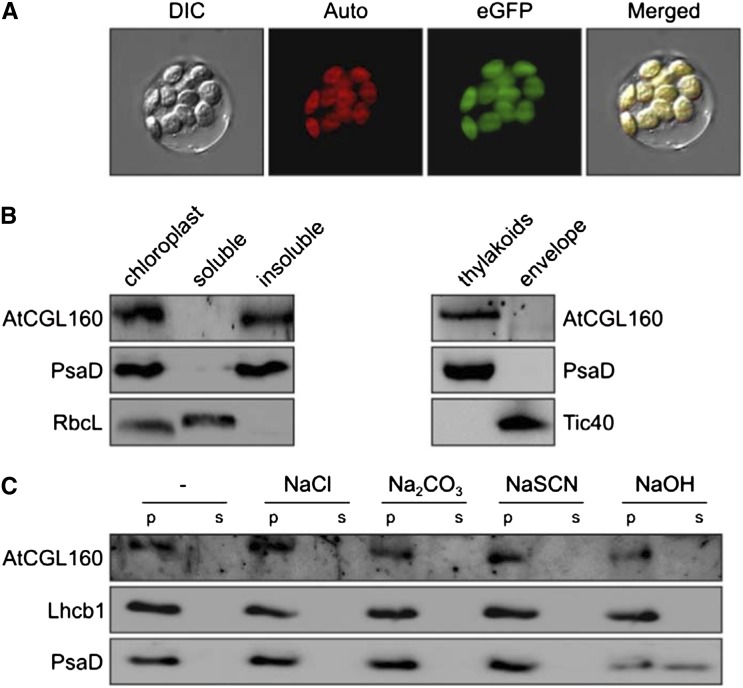
To determine the subcellular localization of AtCGL160, isolated protoplasts from atcgl160-1 plants overexpressing AtCGL160-eGFP were analyzed. The eGFP fluorescence signals were detected exclusively in chloroplasts (Fig. 5A), as expected given the chloroplast location of CrCGL160 in C. reinhardtii (Terashima et al., 2011). To study the suborganellar location of AtCGL160, chloroplasts were fractionated into insoluble and soluble fractions as well as into a thylakoid membrane fraction and a chloroplast envelope fraction (Fig. 5B). Immunoblot analyses with antibodies raised against AtCGL160, and antibodies specifically recognizing marker proteins of chloroplast subcompartments as controls, showed that AtCGL160 is present in the insoluble and thylakoid membrane fractions but not in the envelope. To clarify whether AtCGL160 is an integral or peripheral thylakoid protein, thylakoids from wild-type (Col-0) plants were treated with alkaline and chaotropic salts to release membrane-associated proteins (Fig. 5C). In this assay, AtCGL160 behaved like the integral protein Lhcb1 rather than the peripheral PsaD1, indicating that it is an integral membrane protein, as already suggested by its four predicted TMs (Fig. 1).
Figure 5.
AtCGL160 is an intrinsic thylakoid protein. A, Subcellular localization of AtCGL160-eGFP. Protoplasts were isolated from oeAtCGL160-eGFP lines as described in “Materials and Methods.” Protoplast integrity was examined by bright-field microscopy. Signals derived from AtCGL160-eGFP (eGFP) and Chl autofluorescence (Auto) were merged with the bright-field image (DIC, for differential interference contrast) to yield the image (Merged) on the right. B, Suborganellar localization of AtCGL160. Intact chloroplasts were isolated from wild-type (Col-0) plants and fractionated into a soluble and an insoluble portion, or a thylakoid and an envelope fraction, as described in “Materials and Methods.” Fractions were analyzed for purity by immunolabeling of the extrinsic thylakoid protein PsaD and the soluble (stromal) large subunit of Rubisco (RbcL). TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS40 (Tic40) served as an envelope-specific marker protein. C, Extraction of thylakoid-associated proteins with chaotropic salt solutions or alkaline pH. Thylakoid membranes from wild-type (Col-0) plants were resuspended at 0.5 mg Chl mL−1 in 10 mm HEPES-KOH, pH 7.5, containing 2 m NaCl, 0.1 m Na2CO3, 2 m NaSCN, 0.1 m NaOH, or no additive. After incubation for 30 min on ice, supernatants containing the extracted proteins (s) and membrane fractions (p) were fractionated by SDS-PAGE and immunolabeled with antibodies raised against AtCGL160, PsaD (as a control for peripheral membrane proteins), or Lhcb1 (as a control for integral membrane proteins).
AtCGL160 Is Not a Subunit of the cpATPase
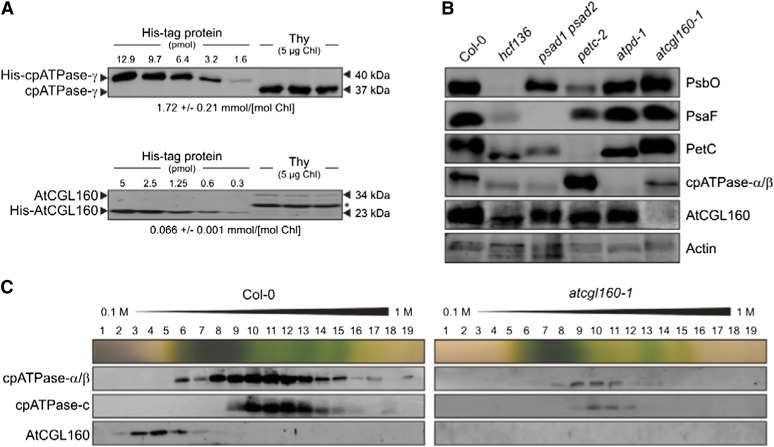
To determine the stoichiometry of AtCGL160 with respect to the cpATPase, signals obtained from known amounts of heterologously expressed and purified AtCGL160 and the γ-subunit of the cpATPase after immunolabeling with appropriate antibodies were compared with those from wild-type (Col-0) thylakoid samples (Fig. 6A). The level of cpATPase-γ was approximately 1.7 mmol mol−1 Chl, or about 80% higher than was found previously in spinach (Kirchhoff et al., 2002). The corresponding value for AtCGL160 was only approximately 0.07 mmol mol−1 Chl. Hence, the ratio of the cpATPase complex to AtCGL160 is about approximately 25:1.
Figure 6.
AtCGL160 is not a subunit of the major thylakoid multiprotein complexes. A, Absolute abundance of AtCGL160 proteins in wild-type (Col-0) plants. Mature AtCGL160 without its membrane domain was expressed in E. coli as a C-terminal fusion to the His6 tag, purified by affinity chromatography, and quantified. Appropriate quantities of His-AtCGL160, as well as His6 fused to cpATPase-γ, were titrated against thylakoid membrane protein preparations and subjected to immunoblot analyses. Representative results from five experiments are shown. The calculated concentrations of AtCGL160 and cpATPase-γ are given below the gels. The asterisk marks an unspecific signal. The molecular masses of proteins are indicated on the right. B, Immunoblot analysis of leaf proteins (corresponding to 40 µg of total protein) from the wild type (Col-0), atcgl160-1, and mutants devoid of PSII (hcf136), PSI (psad1 psad2), Cyt b6f (petc-2), or cpATPase (atpd-1). Antibodies specific for AtCGL160 and representative subunits of the respective thylakoid multiprotein complexes, or actin as a control, were used. C, Analysis of AtCGL160-containing complexes by Suc gradient sedimentation. Thylakoids (1 mg Chl mL−1) from wild-type (Col-0) and mutant (atcgl160-1) plants were solubilized with 1% (w/v) β-DM and separated by centrifugation in a linear 0.1 to 1 m Suc gradient. Nineteen fractions were collected (numbered from top to bottom), and proteins were precipitated from each fraction, separated by SDS-PAGE, blotted onto PVDF membranes, and detected with antibodies raised against the cpATPase subunit α/β or c or against AtCGL160. An image of the Suc gradient is shown above each set of blots.
In addition, the accumulation of AtCGL160 was analyzed in various mutant lines devoid of PSII (high chlorophyll fluorescence136 [hcf136]), PSI (psad1 psad2), Cyt b6f (petc-2), or the cpATPase complex (atpd-1; Fig. 6B). The absence of each complex was verified by immunological screening for marker proteins (PsbO for PSII, PsaF for PSI, PetC for Cyt b6f, and cpATPase-α/β for cpATPase). As expected, signals for PsbO, PsaF, PetC, and cpATPase-α/β could not be detected in the hcf136, psad1 psad2, petc-2, and atpd-1 lines, respectively. AtCGL160 was present in all mutants except atcgl160-1. Hence, AtCGL160 accumulation does not depend on the presence of the cpATPase and is also independent of the integrity of the other thylakoid multiprotein complexes examined.
In order to ascertain whether AtCGL160 is stably associated with the cpATPase, solubilized wild-type (Col-0) and atcgl160-1 thylakoid membranes were fractionated on Suc gradients, and comigration of AtCGL160 with the cpATPase was examined by immunodetection (Fig. 6C). As expected (Fig. 4), the cpATPase subunits α/β and c were less abundant in atcgl160-1 than in wild-type Suc fractions, and no AtCGL160 was detected in atcgl160-1 Suc fractions. Whereas ATPase-α/β and ATPase-c signals were most prominent in fractions 9 to 13, AtCGL160 accumulated mainly in fractions 3 to 6, making it unlikely that AtCGL160 forms a stable complex with the cpATPase.
Thus, three findings argue that AtCGL160 does not associate stably with the cpATPase: (1) it is some 25 times less abundant than the cpATPase; (2) it accumulates even in the absence of the cpATPase; and (3) it does not comigrate with the complex in Suc gradients.
Loss of AtCGL160 Alters the Transcription of Chloroplast Genes for cpATPase Subunits and Induces a General Decrease in Chloroplast Protein Synthesis
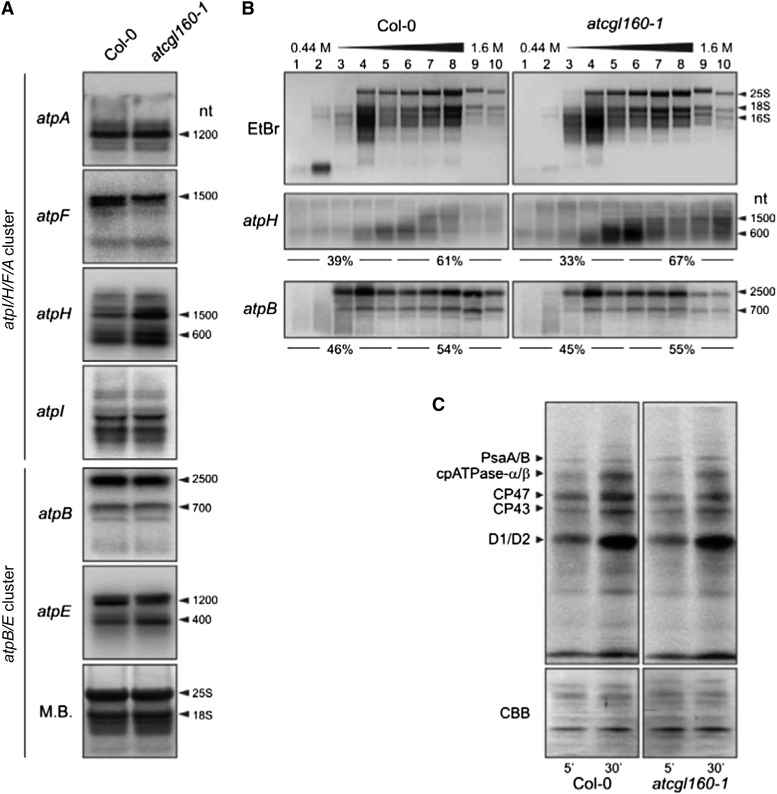
One possible explanation for the reduction in cpATPase levels seen in atcgl160-1 is that the accumulation or processing of transcripts of the chloroplast genes that code for cpATPase subunits is disrupted. Of the nine cpATPase subunits, six are encoded in the chloroplast by genes that were disposed in two gene clusters. The larger atpI/H/F/A gene cluster encodes cpATPase-a, cpATPase-c, cpATPase-b, and cpATPase-α and the small atpB/E gene cluster codes for cpATPase-β and cpATPase-ε. The large gene cluster includes several promoters and gives rise to polycistronic precursors that are processed into at least 18 different transcripts, as shown in maize (Zea mays; Pfalz et al., 2009), whereas two dicistronic atpB/E RNAs and one monocistronic atpE transcript are generated by initiation at two different promoters upstream of the atpB/E coding region and one promoter internal to atpB, respectively (Ghulam et al., 2012). RNA-blot analyses with probes specific for the small gene cluster (for primers and binding information, see Supplemental Table S2) revealed no changes in atpB transcript patterns or amounts. However, for atpE, a 1,200-nucleotide fragment was less abundant in atcgl160-1, while a 400-nucleotide fragment was overrepresented relative to the wild type (Col-0; Fig. 7A). Some differences were also observed in the output of the large gene cluster. RNA-blot analyses of atpA and atpI revealed no alterations, but three atpH transcripts were found to be relatively more abundant in atcgl160-1 than in the wild type. In addition, the pattern of atpF transcripts in atcgl160-1 differed from that in the wild type, with one species being almost completely absent. Overall, the moderate effects on transcript accumulation/processing associated with the loss of AtCGL160 seem unlikely to account for the drastically reduced cpATPase accumulation in atcgl160-1, in striking contrast to the effects seen in the maize pentatricopeptide repeat protein10 mutant, where severe changes in transcript processing result in dramatic alterations in thylakoid protein accumulation (Pfalz et al., 2009).
Figure 7.
Transcriptional and translational profiling of chloroplast-encoded cpATPase subunits. A, RNA-blot analyses of chloroplast-encoded cpATPase genes. Total RNA was isolated from wild-type (Col-0) and atcgl160-1 leaves, and 5-μg aliquots were fractionated on denaturing RNA gels, blotted onto nylon membranes, and hybridized with 32P-labeled probes specific for the indicated transcripts (for primer information, see Supplemental Table S2). To control for loading, blots were stained with methylene blue (M.B.). Signals were detected by the Typhoon system. B, Association of atpH and atpB mRNAs with polysomes. Whole-cell extracts from Col-0 and atcgl160-1 plants were fractionated in linear 0.44 to 1.6 m (15%–55%) Suc gradients by ultracentrifugation. Gradients were divided into 10 fractions, and RNA was isolated from equal volumes. RNA gels were stained with ethidium bromide (EtBr) to visualize the distribution of cytosolic (25S and 18S) and chloroplastic (16S) rRNAs and then hybridized with probes specific for atpH and atpB. Signals for atpH (corresponding to the 600-nucleotide [nt] band) and atpB (corresponding to the 2,500-nucleotide band) were quantified (see “Materials and Methods”), and relative values for fractions 1 to 5 and fractions 6 to 10 were calculated and are given below the atpH and atpB gels. C, Pulse-labeling analyses of wild-type (Col-0) and atcgl160-1 leaf discs. Cytosolic translation was inhibited by infiltrating leaves with cycloheximide. Then, leaves were exposed to a flux of 100 µmol photons m−2 s−1 in the presence of [35S]Met. After pulse labeling for 5 and 30 min, samples were washed and thylakoid membranes were fractionated on SDS-PAGE gels. After blotting, newly synthesized proteins were detected with a Storage Phosphor Screen (Fuji) and the Typhoon Phosphor Imager (GE Healthcare). Blotted proteins were visualized by Coomassie Brilliant Blue G-250 (CBB) staining of the membrane to control for equal loading.
To explore a possible role of AtCGL160 in the translation of chloroplast-encoded cpATPase subunits, polysome analyses and pulse-labeling experiments were performed (Fig. 7, B and C). Ribosome-transcript association can be examined by the fractionation of polysome-enriched samples on Suc gradients (Fig. 7B). If translation proceeds normally, multiple ribosomes associate with each transcript, forming polysomes that migrate farther into the gradient (Fig. 7B, fractions 6–10). Low rates of translation initiation or elongation correlate with less efficient ribosome loading, and free ribosomes are enriched in the less dense fractions toward the top of the gradient (Fig. 7B, fractions 1–5). The distribution of ribosomes in the gradient was visualized by staining the ribosomal RNAs (rRNAs) isolated from each fraction with methylene blue after electrophoresis on agarose gels. Cytosolic (25S and 18S) and chloroplastic (16S) rRNA distributions in atcgl160-1 fractions did not differ from those in wild-type fractions, suggesting that lack of AtCGL160 has little or no effect on translational capacity. Ribosome loading on atpH and atpB transcripts (as markers for the atpI/H/F/A and atpB/E operons, respectively) was examined by RNA-blot analyses. Total amounts of atpH transcripts in atcgl160-1 Suc gradients were higher, as expected (Fig. 7A), and a disproportionate accumulation of atpH in fraction 10 was observed. In addition, a selective depletion of atpB transcripts in fractions 9 and 10 from the cgl160-1 sample could be observed. Nonetheless, the ratio of ribosome-associated transcripts to free transcripts (67%/33% = approximately 2) was close to that in the wild type (61%/39% = approximately 1.6; Fig. 7B). The same behavior was noted for atpB; the ratio of polysome-bound to free transcripts in atcgl160-1 (55%/45% = approximately 1.2) was as high as in the wild type (54%/46% = approximately 1.2). Hence, the absence of AtCGL160 does not lead to drastic changes in ribosome loading indicative of defects in translation or translation initiation (Schult et al., 2007; Rott et al., 2011).
To determine whether the reduced accumulation of cpATPase in atcgl160-1 is due to a more general fall in rates of protein synthesis in chloroplasts, in vivo pulse-labeling experiments were performed (Fig. 7C). After inhibition of cytosolic translation with cycloheximide, synthesis rates were examined by radiolabeling newly synthesized chloroplast-encoded proteins with [35S]Met. Signals were assigned to PsaA/B, ATPase-α/β, CP47, CP43, and D1/D2 subunits according to Armbruster et al. (2010). In atcgl160-1 thylakoids, newly synthesized PsaA/B, ATPase-α/β, CP47, and CP43 subunits were less abundant than in wild-type fractions after 30 min, as were D1/D2 after 5 min. ATPase-α/β synthesis rates in atcgl160-1 chloroplasts, calculated on the basis of signal quantification after pulse labeling for 5 and 30 min, were reduced to about 70% of wild-type rates. Hence, AtCGL160 disruption leads to a moderate overall reduction in chloroplast protein synthesis, presumably because supplies of ATP in atcgl160-1 are insufficient to support normal rates of translational elongation.
AtCGL160 Is Involved in Integration of the c-Subunit into the cpATPase Complex
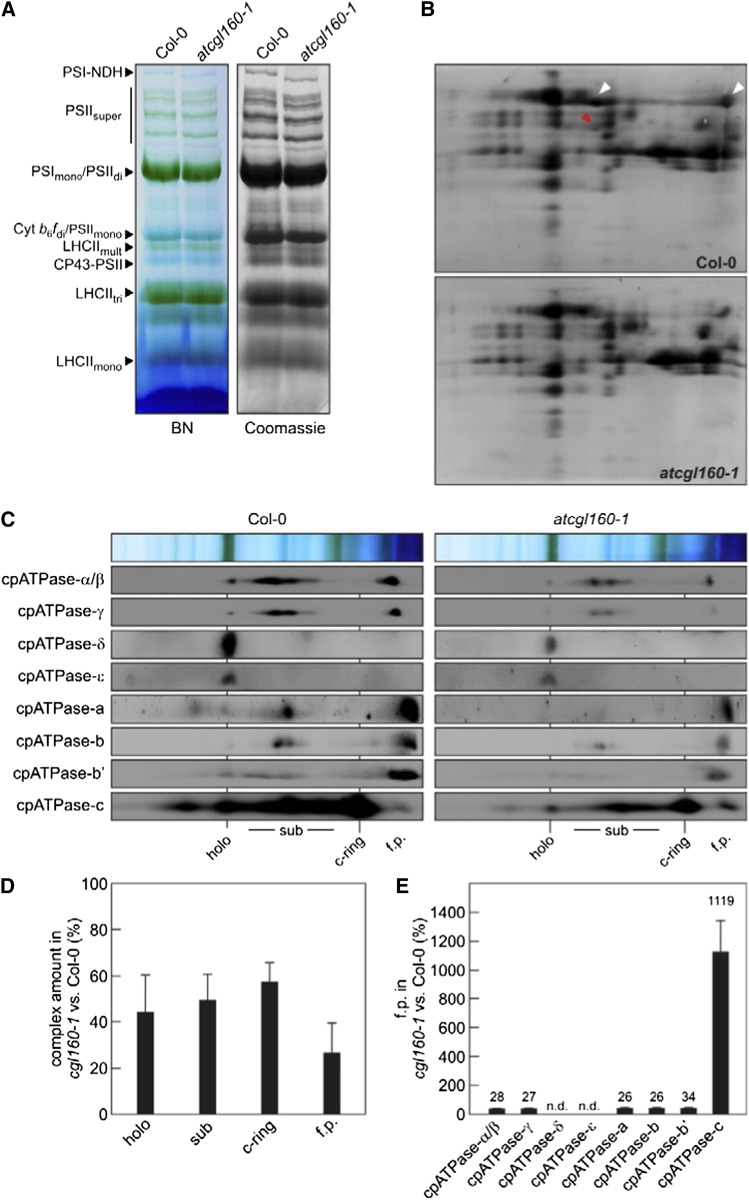
To assess the impact of AtCGL160 on cpATPase assembly, we performed comparative blue native (BN)/SDS-PAGE analyses. Intact thylakoid membrane complexes were isolated and solubilized with n-dodecyl β-d-maltoside (β-DM), and proteins were then fractionated in the presence of Coomassie Brilliant Blue G-250 (Fig. 8A). Complexes were identified according to Armbruster et al. (2010). The complexes resolved by BN-PAGE were then separated into their subunits in the second dimension by electrophoresis on SDS-PAGE gels and blotted onto polyvinylidene difluoride (PVDF) membranes. The cpATPase subunits α/β and γ were readily visible after Coomassie blue staining of the membrane (Fig. 8B) and were less abundant in atcgl160-1, as expected from our immunoblot (Fig. 4) and Suc gradient (Fig. 6C) analyses. The formation of the cpATPase complex was then examined by immunodetection of individual cpATPase subunits after two-dimensional fractionation of the complexes by BN-PAGE and SDS-PAGE (Fig. 8C). Prominent signals were obtained at several positions in both samples. The largest migrates at a position corresponding to PSI monomers/PSII dimers on the BN gel (with a molecular mass of about 580 kD; Fagioni et al., 2009), contains all cpATPase subunits, and thus represents the functional cpATPase holocomplex (calculated molecular mass of 578.6 kD; designated as “holo” in Fig. 8C). Several other signals from individual subunits were obtained at positions between those of PSI monomers/PSII dimers (580 kD) and LHCII trimers (155 kD) and should represent subcomplexes. Protein profiles obtained previously by BN-PAGE and liquid chromatography-tandem mass spectrometry (Takabayashi et al., 2013) have indicated that one stable subcomplex in the assembly of the cpATPase might correspond to CF1 with the δ-subunit (α3β3γε; calculated molecular mass of 378.6 kD); however, in our study, only α-, β-, and γ-subunits from CF1 and a- and b-subunits from CFo could be detected at similar positions (Fig. 8C). The smallest subcomplex found below LHCII trimers (155 kD) appears to represent the c-ring from CFo (calculated molecular mass of 110.6 kD; Fig. 8C).
Figure 8.
Accumulation of cpATPase complexes. A, BN analysis of thylakoid multiprotein complexes from wild-type (Col-0) and atcgl160-1 plants. Thylakoid extracts (80 μg of Chl) were solubilized with the mild detergent β-DM (1% [w/v]) and fractionated by BN-PAGE. Complexes containing no Chl-binding proteins were visualized by staining the gel with Coomassie Brilliant Blue G-250. The bands detected were assigned to specific protein complexes according to Armbruster et al. (2010): the PSI-NAD(P)H dehydrogenase supercomplex (PSI-NDH), PSII supercomplexes (PSIIsuper), PSI monomers and PSII dimers (PSImono and PSIIdi), PSII monomers and dimeric Cyt b6f (PSIImono and Cyt b6/fdi), multimeric LHCII (LHCIImult), CP43-free PSII monomers (CP43-PSII), trimeric LHCII (LHCIItri), and monomeric LHCII (LHCIImono). B, Two-dimensional BN/SDS-PAGE separation of thylakoid protein complexes. Individual lanes from BN-PAGE gels as in A were analyzed in the presence of SDS by electrophoresis on Tris-Tricine gels (10%, 4 m urea) and staining with colloidal Coomassie Brilliant Blue G-250. The identities of the α/β-subunits (white arrowheads) and the γ-subunit (red arrowhead) of cpATPase are indicated on the Col-0 gel. C, Detection of cpATPase subcomplexes by immunoblot analyses of two-dimensional BN/SDS gels as in B, employing antibodies specific for individual cpATPase subunits. The positions of the fully assembled cpATPase (holo), cpATPase subcomplexes (sub), the cpATPase c-ring (c-ring), and free proteins (f.p.) are indicated. D, Quantification of holo, sub, c-ring, and f.p. Holo, sub, and f.p. amounts were quantified based on α/β and γ immunodetection assays. Chemiluminescence signals for atcgl160-1 and wild-type samples assessed in parallel experiments were quantified as described in “Materials and Methods.” Levels of atcgl160-1 signals relative to wild-type signals are shown as percentages. Averages of holo, sub, and f.p. were calculated from nine independent experiments, and averages of c-ring ratios were determined from four independent experiments. Error bars reflect sd. E, Monomers of α/β-, γ-, a-, b-, b′-, and c-subunits were quantified from immunodetection assays as described in “Materials and Methods.” Levels of chemiluminescence signals detected for monomers in cgl160-1 samples relative to those detected in wild-type samples are given as percentages. Values are averages of three to five independent measurements, and error bars indicate sd.
After quantification of the signals for the holocomplex and the larger subcomplexes in Figure 8C, it became clear that there is an overall deficiency of cpATPase subunits in atcgl160-1 (Fig. 8D), such that the holocomplex, subcomplex, and c-ring were reduced to about 40% to 60% of their levels in wild-type plants (Fig. 8D).
In wild-type and atcgl160-1 thylakoids, α-, β-, γ-, a-, b-, b′-, and c-subunits were also detectable in their free, monomeric forms. Difference ratios (atcgl160-1:wild type) for all monomers were calculated, and the levels of almost all of these subunits were found to be reduced relative to the wild type, by between approximately 3-fold (to 34% of the wild-type level for b′) and approximately 4-fold (to about 25% of the wild-type levels for α, β, γ, a, and b). Most strikingly, 11 times more free c-subunits were detected in the monomeric state in atcgl160-1 extracts than in wild-type plants (Fig. 8E). This clearly indicates that the assembly of the c-subunits into the cpATPase complex is specifically disrupted in the atcgl160-1 mutant.
AtCGL160 Interacts Directly with the c-Subunit of the cpATPase
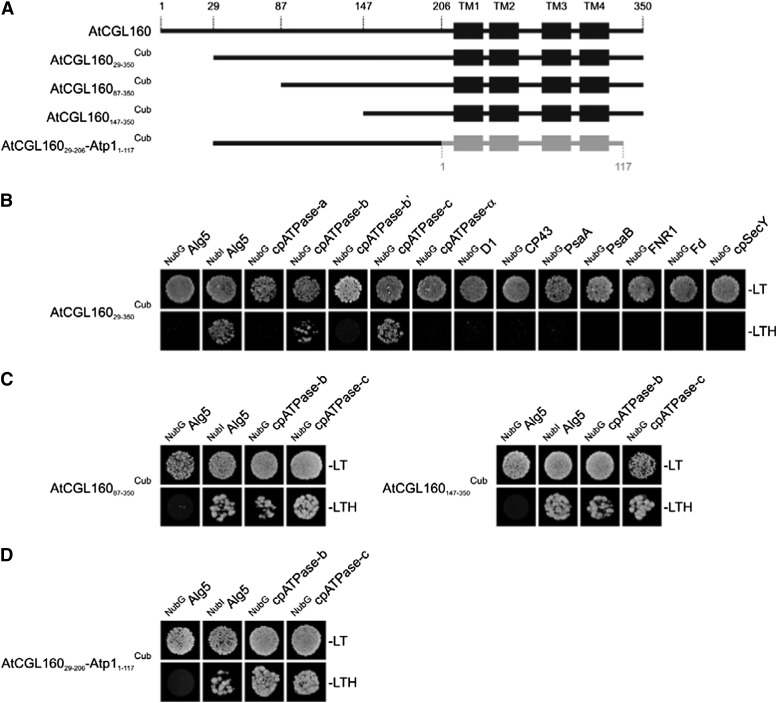
AtCGL160 does not form stable complexes with the cpATPase (Fig. 6C). So to identify transient associations between AtCGL160 and cpATPase subunits, we examined its interaction with other thylakoid proteins using the split-ubiquitin system (Fig. 9). In this assay, the mature form of AtCGL160 did not interact with selected subunits of PSII (D1 and CP43) or of PSI (PsaA and PsaB) or with proteins like ferredoxin-NADP+ oxidoreductase (FNR), ferredoxin (Fd), and cpSecY (a component of the secretory thylakoid targeting pathway; Fig. 9B). We then tested for the interaction of AtCGL160 with those components of the CFo subcomplex that were most severely affected by the lack of AtCGL160 (subunits a, b, b′, and c) as well as with the α-subunit from CF1 as a control. Whereas no interaction with the α-, a-, or b′-subunit of the cpATPase was detectable, AtCGL160 clearly interacted with the ring-forming c-subunit and, to a certain degree, also with the b-subunit in split-ubiquitin assays. To delineate the interaction domain, N-terminally truncated AtCGL160 constructs were also tested in the split-ubiquitin system (Fig. 9, A and C). AtCGL16087-350Cub and AtCGL160147-350Cub, which lack 86 and 146 amino acids at the N terminus, respectively, were still capable of interacting with the b- and c-subunits of CFo. Because neither the N-terminal domain (amino acids 29–206) nor the predicted membrane domain (amino acids 207–350) of AtCGL160 alone, when fused to the Cub domain, interacted with NubIAlg5 in positive control experiments, the interactions of these domains with the c-subunit could not be tested. Because the C-terminal half of AtCGL160 is related to the atp1/uncI gene product of Synechocystis sp. PCC6803, we also analyzed interactions of Atp1 with cpATPase subunits b and c of Arabidopsis. Atp1 alone (Atp11-117Cub) failed to interact with NubIAlg5 in positive control experiments and, therefore, could not tested for its capacity to interact with the c-subunit. Interestingly, however, when Atp1 was fused C terminally to amino acids 29 to 206 of AtCGL160, the hybrid interacted with subunits b and c as efficiently as did the AtCGL16029-350 construct.
Figure 9.
Split-ubiquitin assays for interactions between AtCGL160 and selected thylakoid proteins. A, Schematic depictions of the four constructs used in the tests presented in B to D. TM domains of AtCGL160 and Atp1 are schematically shown as black and gray boxes, respectively, and amino acid positions are indicated above the full-length sequence of AtCGL160. B, Split-ubiquitin assays with mature AtCGL160 (AtCGL16029-350Cub). Assays were performed using fusions to the C-terminal (Cub) and N-terminal (NubG) halves of ubiquitin. NubIAlg5 (the unrelated endoplasmic reticulum membrane protein Alg5 fused to the wild-type Nub) served as a positive control. Alg5 fused to NubG (NubGAlg5) was used as the negative control. To test for interactions involving the AtCGL160 protein, the mature AtCGL160 protein was fused to Cub (AtCGL160Cub) and the selected thylakoid proteins were fused to NubG. Yeast colonies were first plated on permissive medium (−LT, lacking Leu and Trp; top row) and then on selective medium (−LTH, lacking Leu, Trp, and His; bottom row; see “Materials and Methods”). C, Interaction assays with the two N-terminally truncated fragments AtCGL16087-350 and AtCGL160147-350. D, Split-ubiquitin analyses with a fusion construct consisting of the N terminus of AtCGL160 (amino acids 29–206) and the full-length Atp1 (amino acids 1–117) from Synechocystis sp. PCC6803.
Thus, these experiments show (1) that AtCGL160 interacts with subunits b and c of CFo, (2) that its N-terminal half (amino acids 1–147) is dispensable for these interactions, and (3) that its membrane domain can be functionally replaced by its bacterial counterpart from Synechocystis sp. PCC6803. Together with the finding that the assembly of c-subunits is significantly disrupted in atcgl160-1, this strongly suggests that AtCGL160 is specifically involved in the assembly of the c-ring during cpATPase biogenesis.
DISCUSSION
Lack of AtCGL160 Elicits a Characteristic Low-cpATPase Phenotype
In Arabidopsis, the effects of knockout or knockdown of some of the structural subunits of cpATPase on photosynthesis have been studied in detail. Absence of cpATPase-δ (in atpd-1) or cpATPase-γ (in deficiency of plastid atp synthase1 [dpa1]) has been found to destabilize the entire cpATPase complex, causing seedling lethality (Maiwald et al., 2003; Dal Bosco et al., 2004). In these mutants, residual accumulation of Cyt b6f and of the photosystems still allows linear electron flow. Because of the block in photophosphorylation, atpd-1 and dpa1 display increased NPQ and greater deepoxidation of xanthophyll cycle pigments under low light, which is compatible with a slower dissipation of the transthylakoid proton gradient (Maiwald et al., 2003; Dal Bosco et al., 2004). More recently, cpATPase accumulation was systematically decreased using antisense RNA directed against the nuclear AtpC gene and by the introduction of point mutations into the translation initiation codon of the plastid-encoded atpB gene via chloroplast transformation (Rott et al., 2011). In the resulting lines, cpATPase levels ranging from 100% to less than 10% of the wild-type level were observed. Also in these lines, NPQ was triggered at very low light intensities. Moreover, other evidence suggested that this is due to an increase in the steady-state proton motive force, resulting in strong lumen overacidification (Rott et al., 2011).
The phenotype of plants lacking AtCGL160 is characterized by a high-qE phenotype that becomes manifest even under low light (Fig. 3D) and is compatible with the higher xanthophyll cycle activity revealed by the dramatic increase in zeaxanthin and antheraxanthin levels in atcgl160-1 (Table I). Therefore, the atcgl160-1 phenotype is very similar to the one described by Rott et al. (2011) for lines with decreased cpATPase content and can be entirely attributed to the relative deficiency of cpATPase (as low as 10% of wild-type levels for some CFo subunits; Fig. 4) associated with the absence of AtCGL160.
AtCGL160 Is Involved in c-Ring Assembly
Lack of AtCGL160 leads to moderate changes in the transcription of chloroplast genes for cpATPase subunits, has only weak effects on polysome loading, and causes a general decrease in chloroplast protein synthesis (Fig. 7). Obviously, these effects are not sufficient to explain the “low-cpATPase” phenotype and are probably due to a scarcity of ATP. Thus, the up-regulation of atpH transcription could be a compensatory mechanism activated by impaired cpATPase assembly.
The abundance profiles for the nine cpATPase subunits provide decisive clues to the underlying cause of the effects of AtCGL160 loss on cpATPase function. The Fo subunits a, b′, and c are more severely affected (dropping to approximately 10% of wild-type levels) than the F1 subunits, in particular α, β, and δ (approximately 30% of wild-type levels; Fig. 4), clearly pointing to a primary defect in the Fo complex as the cause for the impairment in cpATPase accumulation. Interestingly, CF1 accumulates in 3-fold excess relative to CFo in atcgl160 thylakoids. This can only be explained by a thylakoid association of CF1 independent of CFo. Finally, the specific increase in the c-monomer fraction, and the interaction of AtCGL160 with the c-subunit in the split-ubiquitin assay, demonstrate that AtCGL160 is required specifically for efficient assembly of the c-ring. Thus, if AtCGL160 is inactivated, the number of c-rings formed is only sufficient for the assembly of approximately 10% of the normal level of Fo subcomplexes, thus accounting for the typical low-cpATPase effects (see above). How c-ring assembly is promoted by AtCGL160 at the molecular level needs to be addressed in future experiments, but it is tempting to speculate that it acts like the related prokaryotic Atp1/UncI proteins. In fact, Ozaki et al. (2008) showed that when the P. modestum c-subunit alone was synthesized in vitro in the presence of liposomes, it failed to form c-rings. However, when the c-subunit was synthesized together with P. modestum UncI, the c-ring was formed. Moreover, fusion of two kinds of liposomes, one containing only unassembled c-subunit and the other only UncI, resulted in gradual formation of the c-ring, establishing that UncI itself can mediate the posttranslational assembly of c-rings in vitro (Ozaki et al., 2008). Our split-ubiquitin assay data, which show that the membrane domain of AtCGL160 can be replaced by Synechocystis sp. PCC6803 Atp1 in its interactions with cpATPase subunits, strongly support the idea that the plant and cyanobacterial proteins have similar functions.
Evolutionary Considerations: AtCGL160 Versus Atp1/UncI Proteins
In prokaryotes, the strongest evidence for an involvement of Atp1/UncI proteins in ATP synthase biogenesis comes from studies of artificial F1Fo-ATP synthases and in vitro assays. Thus, a hybrid F1Fo (F1 from Bacillus PS3 and Fo from P. modestum) expressed in E. coli requires Atp1/UncI from P. modestum for c-ring formation and coupled ATPase activity (Suzuki et al., 2007). Similarly, functional production of the Na+ F1Fo-ATP synthase from A. woodii in E. coli requires the A. woodii atp1/uncI gene for proper assembly (Brandt et al., 2013). In addition, c-subunit monomers and c-rings copurify together with P. modestum UncI/Atp1 (Suzuki et al., 2007), and oligomerization of P. modestum c-subunits into c11-rings is mediated by Atp1/UncI in vitro (Ozaki et al., 2008). Nevertheless, deletion of prokaryotic atp1/uncI genes has only modest effects on the corresponding mutant strains. Thus, deletion of uncI in E. coli merely results in a slightly reduced growth yield (Gay, 1984). Likewise, atp1 deletion mutants of B. pseudofirmus OF4 synthesize normal amounts of active ATP synthase and its subunits (Liu et al., 2013). However, an in-depth analysis showed that in the B. pseudofirmus atp1 deletion mutant the c-rotor is less stable than in wild-type plants, as indicated by the presence of free c-monomers in the former (Liu et al., 2013). However, the decrease in c-ring accumulation due to dissociation was very slight in the mutant line and was not associated with a drop in the levels of the other ATP synthase subunits (Liu et al., 2013), in striking contrast to the effects on cpATPase subunit accumulation in atcgl160-1 plants (Fig. 4).
Why does the lack of Atp1/UncI have such a mild effect on ATP synthase function while AtCGL160 appears to be central to c-ring assembly in the flowering plant Arabidopsis, such that in its absence only 20% of normal levels of functional ATP synthase can accumulate? In principle, two explanations are available. Either AtCGL160 has additional functions in ATP synthase assembly that are perhaps mediated by its N-terminal half, or AtCGL160 and its prokaryotic counterparts perform essentially the same biochemical function and the difference in severity of their mutant phenotypes is due to more general differences in the details of the assembly process between prokaryotes and plants. We favor the second possibility and propose that the function of the eukaryote-specific N-terminal half of AtCGL160 is a regulatory one, as indicated by its capacity to become reversibly phosphorylated (Reiland et al., 2009). Cross-species comparisons have shown that prokaryotes often tolerate mutations in assembly factors, whereas lack of their plant counterparts has severe effects on the accumulation of the respective multiprotein complex. Examples include the cyanobacterial PSII assembly factors Ycf48 and Sll0933 and their Arabidopsis counterparts HCF136 and PHOTOSYNTHESIS AFFECTED MUTANT68 (PAM68; Meurer et al., 1998; Komenda et al., 2008; Armbruster et al., 2010). A general explanation for the more severe phenotype of the Arabidopsis atcgl160-1 mutation compared with prokaryotic mutations (and correspondingly for hcf136 and pam68 in Arabidopsis and ycf48 and sll0933 in Synechocystis sp. PCC6803) might be provided by the more effective elimination of nonassembled proteins and abnormal complexes by the proteolytic quality-control system in chloroplasts (for review, see Nixon et al., 2010).
MATERIALS AND METHODS
Plant Material and Growth Conditions
The atcgl160-1 (SALK_057229; Col-0 background) and atcgl160-2 (WiscDsLoxHs024_02B; Col-0 background) T-DNA lines were obtained from the SALK collection (Alonso et al., 2003) and the Wisconsin DsLox T-DNA collection (Woody et al., 2007), respectively. Arabidopsis (Arabidopsis thaliana) plants were grown on potting soil (A210; Stender) under controlled greenhouse conditions (70–90 µmol photons m−2 s−1, 16-/8-h light/dark cycles) or for biochemical and physiological analyses in climate chambers under short-day conditions (8/16 h of light/dark). When grown under greenhouse conditions, fertilizer was added according to the manufacturer’s recommendations (Osmocote Plus; 15% [w/v] nitrogen, 11% [w/v] P2O5, 13% [w/v] K2O, and 2% MgO [w/v]; Scotts Deutschland). Mutant lines unable to survive photoautotrophically (psad1 psad2, atpd-1, petc-1, and hcf136) were grown in climate chambers (16-/8-h light dark cycles) on Murashige and Skoog agar plates (Duchefa) supplemented with 1.5% (w/v) Suc.
To rescue the wild-type phenotype, the AtCGL160 coding region was cloned into the binary Gateway destination vector pB7FWG2.0 (Karimi et al., 2002), placing the gene under the control of the 35S promoter derived from the Cauliflower mosaic virus and 5′ of the EGFP gene to generate an AtCGL160-eGFP fusion. The construct was first transformed into Agrobacterium tumefaciens strain GV3101 and then into atcgl160-1 plants by the floral dip method (Clough and Bent, 1998). Seeds were collected after 4 weeks, and transformed T1 plants were selected after several BASTA treatments. Expression of the AtCGL160-eGFP construct was assessed by northern-blot and western-blot analyses. Complementation of the atcgl160-1 mutant phenotype was checked by measuring leaf surface areas from 8- to 32-d-old plants grown under short-day conditions and by NPQ analyses (Supplemental Fig. S1).
Chl a Fluorescence Measurements
Chl a fluorescence measurements were carried out with the Dual-PAM 101/103 fluorometer (Walz). Plants were dark adapted for 20 min before the experiments. Single leaves were exposed briefly (30 s) to measuring light for minimal fluorescence (F0) measurement and to a saturating light pulse (10,000 µmol photons m−2 s−1, 800 ms) to determine maximal Chl fluorescence yield (Fm). Actinic red light was switched on for 15 min, and steady-state fluorescence yields (FS) were measured. The maximum fluorescence yield in the light (Fm′) was determined by applying a saturating light pulse (10,000 µmol photons m−2 s−1, 800 ms) at the end of the light phase. After the 10-min dark relaxation phase, a saturating light pulse (10,000 µmol photons m−2 s−1, 800 ms) was administered to measure the maximal fluorescence yield in the dark relaxation phase (Fm″) and the minimal fluorescence in the dark relaxation phase (F0″). Parameters were calculated as follows: minimum fluorescence yield in the light, F0′ = F0/(Fv/Fm + F0/Fm′) (Oxborough and Baker, 1997); maximum quantum yield of PSII, Fv/Fm = (Fm − F0)/Fm (Genty et al., 1989); effective quantum yield of PSII, ΦII = (Fm′ − F0′)/Fm′ = Fv′/Fm′ (Genty et al., 1989); ETR through PSII, ETR(II) = ΦII × PAR × 0.84 × 0.5 (Schreiber et al., 1994); excitation pressure, 1-qP = 1 − (Fm′ − FS)/(Fm′ − F′) (Dietz et al., 1985); NPQ, NPQ = (Fm − Fm′)/Fm′ (Bilger and Björkman, 1990); qE, qE = Fm/Fm′ − Fm/Fm′′ (Thiele et al., 1997); and qI, qI = qIDAS + qILAS = (0.83 − Fv/Fm) + (Fv − Fv′′)/Fv, where qIDAS = photoinhibitory quenching in the dark-adapted state and qILAS = photoinhibitory quenching in the light-adapted state (Björkman and Demmig, 1987).
ETR, NPQ, qE, and qI values were also determined using seven successive slow induction/dark relaxation assays in which the actinic light intensity during the light phase was steadily increased (13, 22, 53, 95, 216, 339, and 531 µmol photons m−2 s−1).
Chl a fluorescence yields of whole plants were recorded using an imaging Chl a fluorometer (Imaging PAM; Walz). After dark adaptation for 20 min, blue measuring light (1 Hz, intensity 4) was switched on, and whole plants were exposed to a first saturating light pulse (intensity 4). A second saturating light pulse was applied at the end of the actinic light phase (10 min, 100 µmol photons m−2 s−1). Y(NPQ) values were calculated as described by Kramer et al. (2004).
Nucleic Acid Analysis
Arabidopsis DNA was isolated from 3-week-old leaves as described by Ihnatowicz et al. (2004). T-DNA insertion sites for atcgl160-1 and atcgl160-2 were determined by PCR using combinations of insertion-specific and gene-specific primers (Supplemental Table S2). Total RNA was extracted from fresh leaves using the TRIzol reagent (Invitrogen). SuperScript III reverse transcriptase (Invitrogen) and dT oligomers were employed for reverse transcription according to the manufacturer’s instructions. Transcript abundance of AtCGL160 was examined by PCR and by real-time PCR using the Bio-Rad IQ5 Multicolor Real-Time PCR Detection System (for primer information, see Supplemental Table S2).
Northern analyses were carried out under stringent conditions according to standard protocols (Sambrook and Russell, 2001). Samples equivalent to 5 µg of total RNA were fractionated by electrophoresis on formaldehyde-containing agarose gels (1.5% [w/v]), blotted onto nylon membranes (Hybond-N+; Amersham Bioscience), and fixed by UV radiation (Stratalinker UV Crosslinker 1800). Methylene blue staining of rRNAs (0.02% [w/v] methylene blue, 0.3 m sodium acetate, pH 5.5) was carried out to control for equal loading. To detect AtCGL160- and cpATPase-specific transcripts, amplified DNA fragments from cDNA were labeled with radioactive [α-32P]dCTP and subsequently used for hybridization (for primer information, see Supplemental Table S2). Signals were detected and quantified with the Typhoon Phosphor Imager System (GE Healthcare) and Bioprofile software (PeqLab), respectively.
Polysome-associated mRNAs were isolated according to Barkan (1993). Leaves of 28-d-old plants (approximately 300 mg) were ground in liquid nitrogen and homogenized in polysome extraction buffer (0.2 m Tris-HCl, pH 9, 0.2 m KCl, 35 mm MgCl2, 25 mm EGTA, 0.2 m Suc, 1% [v/v] Triton X-100, 2% [v/v] polyoxyethylene-10-tridecyl ether, 0.5 mg mL−1 heparin, 100 mm β-mercaptoethanol, 100 µg mL−1 chloramphenicol, and 25 µg mL−1 cycloheximide). Microsomal membranes were solubilized with 0.5% (w/v) sodium deoxycholate and centrifuged at 16,000g for 15 min at 4°C. Supernatants were layered on top of Suc gradients (15%–55%) and fractionated by centrifugation (240,000g) for 65 min at 4°C. RNA was extracted from each of 10 Suc gradient fractions with phenol:chloroform:isoamyl alcohol (25:25:1) and precipitated (95% [v/v] ethanol) at room temperature. Dissolved RNA (10 mm Tris-HCl, 1 mm EDTA, pH 8) was subjected to gel electrophoresis and RNA gel-blot analyses as described above.
Leaf Pigment Analysis
Leaves from 5-week-old plants grown under short-day conditions were harvested 4 h after simulated sunrise and homogenized in liquid nitrogen. The samples were treated with 100% acetone and centrifuged (16,000g) for 20 min at 4°C. Pigment compositions in the supernatants were analyzed as described by Färber et al. (1997).
Thylakoid Membrane Isolation and BN/SDS-PAGE
Leaves from 6-week-old plants grown under short-day conditions were harvested shortly before the onset of the light period, and thylakoid membrane-enriched samples were isolated according to Järvi et al. (2011). Chl concentrations were determined as described by Porra et al. (1989). For immunotitrations, thylakoid membrane pellets were frozen in liquid nitrogen and stored at −80°C or directly resuspended in loading buffer (6 m urea, 50 mm Tris-HCl, pH 6.8, 100 mm dithiothreitol, 2% [w/v] SDS, 10% [w/v] glycerol, and 0.1% [w/v] bromphenol blue). Denaturation for 30 s at 70°C and protein fractionation on Tricine-SDS-PAGE gels (10%–15% gels) were carried out according to Schägger (2006). Immunodetections were performed as described below.
Sample preparation for BN-PAGE was performed as described by Peng et al. (2008) with freshly prepared thylakoids. First, membranes were washed four times in washing buffer (20% [w/v] glycerol, 25 mm BisTris-HCl, pH 7). Then, samples were treated with solubilization buffer (1% [w/v] β-DM, 20% [w/v] glycerol, and 25 mm BisTris, pH 7; adjusted to 1 mL mg−1 Chl) for 10 min on ice. After centrifugation (16,000g, 20 min, 4°C), supernatants were supplemented with one-tenth volume of BN sample buffer (100 mm BisTris-HCl, pH 7, 750 mm ε-aminocaproic acid, and 5% [w/v] Coomassie Brilliant Blue G-250). BN-PAGE gels (4%–12% gradient) were prepared as described by Schägger et al. (1994). Solubilized samples corresponding to 80 µg of Chl were loaded per lane, and gels were run at 4°C overnight. To separate complexes into their subunits, BN-PAGE strips were treated with denaturing buffer (0.5 m Na2CO3, 2% [w/v] SDS, and 0.7% [v/v] β-mercaptoethanol) for 30 min at room temperature and loaded on Tricine-SDS-PAGE gels (10% gels supplemented with 4 m urea), which were prepared according to Schägger (2006). Gels were subsequently subjected to immunoblot analysis with antibodies against cpATPase subunits as described below.
Salt Wash Treatment of Thylakoids
Salt wash treatments were performed as described by Karnauchov et al. (1997). Isolated thylakoid membranes were resuspended to a final Chl concentration of 0.5 mg mL−1 in 50 mm HEPES-KOH, pH 7.5, supplemented with 2 m NaCl, 0.1 m Na2CO3, 2 m NaSCN, or 0.1 m NaOH. Suspensions were incubated for 30 min on ice, and soluble membrane proteins were separated by centrifugation at 10,000g for 10 min at 4°C. Pellets and supernatants were treated with solubilization buffer, fractionated on Tricine-SDS-PAGE gels (10%), and subjected to immunoblot analysis using antibodies specific for AtCGL160 (generated in this study), Lhcb1 (Agrisera), and PsaD (Agrisera).
Suc Gradient Fractionation of Thylakoid Membrane Complexes
Leaves from 6-week-old plants were harvested and homogenized in isolation buffer (25 mm MES, pH 6.5, 330 mm Suc, 5 mm MgCl2, 1.5 mm NaCl, and 100 mm NaF). The homogenate was filtered through double-layered Miracloth (Calbiochem) and centrifuged at 4,500g for 5 min at 4°C. Pelleted thylakoids were washed in 5 mm EDTA, pH 7.8, resuspended in water, and solubilized in 1% (w/v) β-DM for 10 min on ice. Solubilized material was separated by centrifugation at 16,000g for 5 min at 4°C and loaded onto a linear 0.1 to 1 m Suc gradient containing 5 mm Tricine-NaOH, pH 8, and 0.05% (w/v) β-DM. Gradients were subjected to ultracentrifugation (191,000g, 4°C) for 21 h. Proteins from 19 fractions were separated on Tricine-SDS-PAGE gels (10%, 4 m urea) and analyzed by immunoblot with antibodies against cpATPase subunits and AtCGL160 as described below.
Chloroplast and Envelope Isolation
Chloroplast isolation was performed according to Kunst (1998). Briefly, leaves from 4-week-old dark-adapted plants (20 g) were homogenized in 450 mm sorbitol, 20 mm Tricine-KOH, pH 8.4, 10 mm EDTA, 10 mm NaHCO3, and 0.1% (w/v) bovine serum albumin (BSA), filtered through two layers of Miracloth (Calbiochem), and centrifuged at 500g for 6 min at 4°C. Pelleted chloroplasts were resuspended in 800 µL of 0.3 m sorbitol, 20 mm Tricine-KOH, pH 8.4, 2.5 mm EDTA, and 5 mm MgCl2, applied to a two-step Percoll gradient (40%–80% [v/v]), and centrifuged at 6,500g for 20 min at 4°C. Intact chloroplasts were collected from the interface and ruptured in a buffer containing 20 mm HEPES-KOH, pH 7.5, 10 mm EDTA for 30 min on ice. Soluble proteins were separated from the insoluble fraction by centrifugation at 42,000g for 30 min at 4°C.
Envelope and thylakoid membranes were isolated from 4-week-old wild-type plants (300 g of leaves). Chloroplast isolation was performed as described above. Envelope and thylakoid membranes were separated on a step gradient (1.2/1/0.46 m Suc) by centrifugation at 58,000g for 2 h as described by Li et al. (1991). Envelope membranes were collected from the interface between the layers of 0.46 and 1 m Suc, and thylakoid membranes were collected from the pellet. Each fraction was washed several times and resuspended in 10 mm HEPES, 5 mm MgCl2, pH 7.6, buffer and used for immunodetection analyses.
Immunoblot Analyses
Proteins fractionated by gel electrophoresis were transferred to PVDF membranes (Immobilon-P; Millipore) using a semidry blotting apparatus (Bio-Rad) as described in the manufacturer’s instructions. After blocking with TBST (10 mm Tris, pH 8.0, 150 mm NaCl, and 0.1% Tween 20) supplemented with 3% BSA or 5% (w/v) milk, the membranes were incubated with antibodies against subunits of PSI (Agrisera), PSII (Agrisera), Cyt b6f (Agrisera), or cpATPase (obtained from Jörg Meurer, University of Munich) or against AtCGL160 (generated in this study) at 4°C overnight. Rabbit antibodies were generated against the N terminus of AtCGL160, which was heterologously expressed in Escherichia coli. To this end, AtCGL16047-201 was cloned into the pET151 vector (Invitrogen), which leads to an N-terminal fusion with a 6xHis tag. The 6xHis-AtCGL16047-201 protein was purified using nickel-nitrilotriacetic acid agarose (Qiagen) following the manufacturer’s instructions and injected into rabbits for antibody production. Antiserum was employed in dilutions of 1:200 to 1:1,000. Signals were detected by enhanced chemiluminescence (ECL kit; Amersham Bioscience) using an ECL reader system (Fusion FX7; PeqLab) and quantified with Bioprofile software (PeqLab).
Protein Titration
Truncated versions of His-tagged AtCGL160 and cpATPase-γ were heterologously expressed and purified from E. coli strain BL21 (Invitrogen), whose codon usage frequency is optimized for eukaryotic gene expression. The AtCGL160 coding sequence (without the predicted cTP and the membrane domain) and the AtpC1 coding sequence (without the predicted cTP) were cloned into the pET151 vector (Invitrogen). For primer information, see Supplemental Table S2. E. coli cultures were grown at 37°C in Luria-Bertani medium (5 g L−1 yeast extract, 10 g L−1 tryptone, and 10 g L−1 NaCl) supplemented with 100 µg mL−1 carbenicillin, and gene expression was induced by isopropylthio-β-galactoside addition (final concentration of 1 mm). After 4 h, synthesized proteins, carrying a 6xHis tag at their N terminus, were purified under denaturing conditions using a nickel-nitrilotriacetic acid agarose resin (Qiagen) according to the manufacturer’s instructions.
To determine the molar ratio of AtCGL160 to cpATPase-γ in wild-type thylakoid samples, AtCGL160 and cpATPase-γ protein concentrations were quantified employing BSA standards. Then, wild-type thylakoid membrane samples corresponding to 5 µg of Chl were spiked with decreasing amounts of His-AtCGL160 and His-cpATPase-γ and fractionated on Tricine-SDS-PAGE gels. The AtCGL160 and cpATPase-γ in wild-type thylakoid samples were immunodetected and quantified by comparison with the signals obtained for the known amounts of added His-AtCGL160 and His-cpATPase-γ.
In Vivo Translation Assay
Leaf discs (five discs for each time point per genotype) of plants grown under short-day conditions were vacuum infiltrated with pulse-chase buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, and 0.2% [v/v] Tween 20) containing 20 μg mL−1 cycloheximide to block cytosolic translation. After vacuum infiltration and light treatment for 30 min, one sample per genotype was collected. The other samples were vacuum infiltrated again with pulse-chase buffer containing 1 mCi of [35S]Met in order to radiolabel newly translated chloroplast proteins. The infiltrated leaf discs were incubated in the radioactive buffer in the light (100 µmol photons m−2 s−1), and samples were collected after 5 and 30 min of incubation. For thylakoid protein preparation, collected samples were washed, ground up in 20 mm HEPES-KOH, pH 7.5, and 10 mm EDTA, filtered, and centrifuged at 16,000g for 10 min at 4°C. Thylakoid pellets were washed and resuspended in the same buffer. Chl concentration was measured according to Arnon (1949). Proteins were separated on denaturing Tricine-SDS gels (8% acrylamide, 4 m urea), blotted onto PVDF membranes, and exposed to Storage Phosphor Screen (Fuji), and signals were detected with the Typhoon Phosphor Imager (GE Healthcare).
Protoplast Isolation
Leaves from 3-week-old atcgl160-1 plants carrying the oeAtCGL160:eGFP construct were cut into small strips and subjected to vacuum infiltration in a buffer containing 2.6 mg mL−1 Macerozyme (Duchefa), 10 mg mL−1 Cellulase (Duchefa), 10 mm MES, pH 5.7, 20 mm KCl, 0.5 mm mannitol, 10 mm CaCl2, and 1 mg mL−1 BSA. After incubation at room temperature in the dark for 4 h, the mixture was filtered through a nylon membrane (pore size, 72 µm) and centrifuged for 10 min at 50g. The protoplast-containing pellet was resuspended in 8.5 mL of MSC solution (10 mm MES, pH 5.7, 20 mm MgCl2, and 120 mg mL−1 saccharose) and overlaid carefully with 2 mL of MMM solution (10 mm MES, pH 5.7, 10 mm MgCl2, 10 mm MgSO4, and 0.5 m mannitol). After centrifugation at 70g for 10 min, protoplasts floating at the interphase were washed once with MMM solution, centrifuged (50g for 10 min) and resuspended in MMM solution. GFP signals and Chl fluorescence from stably transformed protoplasts were detected using an Axio Imager fluorescence microscope (Zeiss).
Split-Ubiquitin Assay
Transient interactions of membrane-integral proteins were analyzed by using the split-ubiquitin system. The experimental procedure essentially followed that described by Armbruster et al. (2010). AtCGL16029-350, AtCGL16087-350, and AtCGL160147-350 were fused to the N terminus of the Cub domain by cloning the respective coding sequence into the multiple cloning site of the pAMBV4 vector (Dualsystems Biotech; for primer information, see Supplemental Table S2). AtCGL16029-206-Atp11-117 was generated by fusion PCR using overlapping primers (Supplemental Table S2) and cloned into the multiple cloning site of the pAMBV4 vector. Potential interaction partners (cpATPase-a, cpATPase-b, cpATPase-b′, cpATPase-c, cpATPase-α, D1, CP43, PsaA, PsaB, FNR1, Fd, and cpSecY) were fused to the C terminus of the prey protein NubG by ligating the amplified coding sequence into the multiple cloning site of pADSL-Nx (Dualsystems Biotech). Plasmid pAlg5-NubG (Alg5NubG) served as a negative control, whereas pAlg5-NubI (Alg5NubI) was employed as a positive control. pAlg5-NubG encodes a fusion between the endoplasmic reticulum membrane protein Alg5 and NubG, which does not interact with the C-terminal part of ubiquitin (Cub). In contrast, pAlg5-NubI encodes a fusion of the wild-type form of NubI to Alg5, which does interact with Cub. Cotransformation and interaction studies were carried out as described by Pasch et al. (2005) using the Dual-Membrane kit (Dualsystems Biotech). After cotransformation, yeast cells were first grown on synthetic, permissive medium lacking the two amino acids Leu and Trp. Then, growth was tested with the same colonies on selective medium lacking also His.
Bioinformatics Sources
Protein and gene sequences were downloaded from The Arabidopsis Information Resource server (http://www.arabidopsis.org) and the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/). Protein sequence alignments were performed using the Vector NTI software (Invitrogen). cTPs were predicted by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). TM-spanning α-helices were predicted by Aramemnon (http://aramemnon.botanik.uni-koeln.de/) for AtCGL160 and its counterparts in grape (Vitis vinifera), rice (Oryza sativa), and maize (Zea mays) or by SCAMPI (http://scampi.cbr.su.se/) for Picea sitchensis, Selaginella moellendorffii, Physcomitrella patens, C. reinhardtii, and prokaryotic Atp1/UncI sequences. Alignments were formatted using Boxshade (http://www.ch.embnet.org/software/BOX_form.html).
Sequence identifiers for AtCGL160 homologs in eukaryotes are as follows: V. vinifera (GI: 225446613), O. sativa (GI: 125532550), Z. mays (GI: 413950882), Picea sitchensis (GI: 148907731), S. moellendorffii (GI: 302814051), Physcomitrella patens (Pp1s184_139V6.4, PACid:18065191), and C. reinhardtii (GI: 159464014). Sequence identifiers for AtCGL160 homologs in cyanobacteria and other bacteria are as follows: Synechocystis sp. PCC 6803 (sll1321), Synechococcus sp. (GI: 170077363), Cyanothece sp. (GI: 220906906), Anabaena variabilis (GI: 75908831), Gloeocapsa sp. (GI: 434391846), Calothrix sp. (GI: 428298284), Nostoc punctiforme (GI: 186684951), Nostoc sp. (GI: 427707295), P. modestum (GI: 45648), and E. coli (GI: 148132).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AtCGL160-eGFP.1 complements the atcgl160-1 mutation.
Supplemental Table S1. Coregulation of AtCGL160 with photosynthesis-related genes.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Paul Hardy for critical reading of the manuscript.
Glossary
- cpATPase
chloroplast ATP synthase/ATPase
- CFo
chloroplast coupling factor o
- CF1
chloroplast coupling factor 1
- Cyt b6f
cytochrome b6f complex
- UTR
untranslated region
- cTP
chloroplast transit peptide
- TM
transmembrane
- T-DNA
transfer DNA
- cDNA
complementary DNA
- Col-0
Columbia-0
- Chl
chlorophyll
- ETR
electron transport rate
- NPQ
nonphotochemical quenching of chlorophyll a fluorescence
- qE
energy-dependent quenching of chlorophyll fluorescence
- qI
photoinhibitory quenching
- Y(NPQ)
quantum yield of regulated heat dissipation in PSII
- rRNA
ribosomal RNA
- BN
blue native
- β-DM
n-dodecyl β-d-maltoside
- PVDF
polyvinylidene difluoride
- BSA
bovine serum albumin
Footnotes
The online version of this article contains Web-only data.
References
- Ackerman SH, Tzagoloff A. (1990a) Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc Natl Acad Sci USA 87: 4986–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman SH, Tzagoloff A. (1990b) ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem 265: 9952–9959 [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arechaga I, Butler PJ, Walker JE. (2002) Self-assembly of ATP synthase subunit c rings. FEBS Lett 515: 189–193 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al. (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz M, Bals T, Gügel IL, Piotrowski M, Kuhn A, Schünemann D, Soll J, Ankele E. (2009) Alb4 of Arabidopsis promotes assembly and stabilization of a non chlorophyll-binding photosynthetic complex, the CF1CF0-ATP synthase. Mol Plant 2: 1410–1424 [DOI] [PubMed] [Google Scholar]
- Biehl A, Richly E, Noutsos C, Salamini F, Leister D. (2005) Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and co-ordination of nuclear and plastid gene expression. Gene 344: 33–41 [DOI] [PubMed] [Google Scholar]
- Bilger W, Björkman O. (1990) Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynth Res 25: 173–185 [DOI] [PubMed] [Google Scholar]
- Björkman O, Demmig B. (1987) Photon yield of O2 evolution and chlorophyll fluorescence characteristics at 77 K among vascular plants of diverse origins. Planta 170: 489–504 [DOI] [PubMed] [Google Scholar]
- Böttcher B, Gräber P. (2000) The structure of the H+-ATP synthase from chloroplasts and its subcomplexes as revealed by electron microscopy. Biochim Biophys Acta 1458: 404–416 [DOI] [PubMed] [Google Scholar]
- Brandt K, Müller DB, Hoffmann J, Hübert C, Brutschy B, Deckers-Hebestreit G, Müller V. (2013) Functional production of the Na+ F1F(O) ATP synthase from Acetobacterium woodii in Escherichia coli requires the native AtpI. J Bioenerg Biomembr 45: 15–23 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dal Bosco C, Lezhneva L, Biehl A, Leister D, Strotmann H, Wanner G, Meurer J. (2004) Inactivation of the chloroplast ATP synthase gamma subunit results in high non-photochemical fluorescence quenching and altered nuclear gene expression in Arabidopsis thaliana. J Biol Chem 279: 1060–1069 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D. (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Schreiber U, Heber U. (1985) The relationship between the redox state of QA and photosynthesis in leaves at various carbon-dioxide, oxygen and light regimes. Planta 166: 219–226 [DOI] [PubMed] [Google Scholar]
- Drapier D, Rimbault B, Vallon O, Wollman FA, Choquet Y. (2007) Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J 26: 3581–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagioni M, D’Amici GM, Timperio AM, Zolla L. (2009) Proteomic analysis of multiprotein complexes in the thylakoid membrane upon cadmium treatment. J Proteome Res 8: 310–326 [DOI] [PubMed] [Google Scholar]
- Färber A, Young AJ, Ruban AV, Horton P, Jahns P. (1997) Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants: the relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching. Plant Physiol 115: 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay NJ. (1984) Construction and characterization of an Escherichia coli strain with a uncI mutation. J Bacteriol 158: 820–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR. (1989) The relationship between the quantum yield of photosynthetic electron-transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Ghulam MM, Zghidi-Abouzid O, Lambert E, Lerbs-Mache S, Merendino L. (2012) Transcriptional organization of the large and the small ATP synthase operons, atpI/H/F/A and atpB/E, in Arabidopsis thaliana chloroplasts. Plant Mol Biol 79: 259–272 [DOI] [PubMed] [Google Scholar]
- Graham LA, Hill KJ, Stevens TH. (1998) Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J Cell Biol 142: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Karpowicz SJ, Heinnickel M, Dewez D, Hamel B, Dent R, Niyogi KK, Johnson X, Alric J, Wollman FA, et al. (2010) Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth Res 106: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth G, Pohl E. (2001) The structure of the chloroplast F1-ATPase at 3.2 A resolution. J Biol Chem 276: 1345–1352 [DOI] [PubMed] [Google Scholar]
- Ihnatowicz A, Pesaresi P, Varotto C, Richly E, Schneider A, Jahns P, Salamini F, Leister D. (2004) Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J 37: 839–852 [DOI] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro EM. (2011) Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem J 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Jia L, Dienhart MK, Stuart RA. (2007) Oxa1 directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex. Mol Biol Cell 18: 1897–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karnauchov I, Herrmann RG, Klösgen RB. (1997) Transmembrane topology of the Rieske Fe/S protein of the cytochrome b6/f complex from spinach chloroplasts. FEBS Lett 408: 206–210 [DOI] [PubMed] [Google Scholar]
- Karpowicz SJ, Prochnik SE, Grossman AR, Merchant SS. (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem 286: 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H, Mukherjee U, Galla HJ. (2002) Molecular architecture of the thylakoid membrane: lipid diffusion space for plastoquinone. Biochemistry 41: 4872–4882 [DOI] [PubMed] [Google Scholar]
- Kol S, Nouwen N, Driessen AJ. (2008) Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes. J Biol Chem 283: 31269–31273 [DOI] [PubMed] [Google Scholar]
- Komenda J, Nickelsen J, Tichý M, Prásil O, Eichacker LA, Nixon PJ. (2008) The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem 283: 22390–22399 [DOI] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE. (2004) New fluorescence parameters for the determination of qa redox state and excitation energy fluxes. Photosynth Res 79: 209–218 [DOI] [PubMed] [Google Scholar]
- Kunst L. (1998) Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol Biol 82: 43–48 [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L, Vaillier J, Benabdelhak H, Velours J, Slonimski PP, di Rago JP. (2001) Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F1-ATPase in heat stress conditions. J Biol Chem 276: 6789–6796 [DOI] [PubMed] [Google Scholar]
- Leister D, Varotto C, Pesaresi P, Niwergall A, Salamini F. (1999) Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol Biochem 37: 671–678 [Google Scholar]
- Li HM, Moore T, Keegstra K. (1991) Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell 3: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hicks DB, Krulwich TA. (2013) Roles of AtpI and two YidC-type proteins from alkaliphilic Bacillus pseudofirmus OF4 in ATP synthase assembly and nonfermentative growth. J Bacteriol 195: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiwald D, Dietzmann A, Jahns P, Pesaresi P, Joliot P, Joliot A, Levin JZ, Salamini F, Leister D. (2003) Knock-out of the genes coding for the Rieske protein and the ATP-synthase δ-subunit of Arabidopsis: effects on photosynthesis, thylakoid protein composition, and nuclear chloroplast gene expression. Plant Physiol 133: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P. (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J. (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot (Lond) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno D, Iino R, Noji H. (2011) Rotation and structure of FoF1-ATP synthase. J Biochem 149: 655–664 [DOI] [PubMed] [Google Scholar]
- Osman C, Wilmes C, Tatsuta T, Langer T. (2007) Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol Biol Cell 18: 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxborough K, Baker NR. (1997) Resolving chlorophyll a fluorescence images of photosynthetic efficiency into photochemical and non-photochemical components: calculation of qP and Fv'/Fm' without measuring Fo'. Photosynth Res 54: 135–142 [Google Scholar]
- Ozaki Y, Suzuki T, Kuruma Y, Ueda T, Yoshida M. (2008) UncI protein can mediate ring-assembly of c-subunits of FoF1-ATP synthase in vitro. Biochem Biophys Res Commun 367: 663–666 [DOI] [PubMed] [Google Scholar]
- Pasch JC, Nickelsen J, Schünemann D. (2005) The yeast split-ubiquitin system to study chloroplast membrane protein interactions. Appl Microbiol Biotechnol 69: 440–447 [DOI] [PubMed] [Google Scholar]
- Peng L, Shimizu H, Shikanai T. (2008) The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J Biol Chem 283: 34873–34879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoryelov D, Yildiz O, Faraldo-Gómez JD, Meier T. (2009) High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat Struct Mol Biol 16: 1068–1073 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Rak M, Zeng X, Brière JJ, Tzagoloff A. (2009) Assembly of F0 in Saccharomyces cerevisiae. Biochim Biophys Acta 1793: 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott M, Martins NF, Thiele W, Lein W, Bock R, Kramer DM, Schöttler MA. (2011) ATP synthase repression in tobacco restricts photosynthetic electron transport, CO2 assimilation, and plant growth by overacidification of the thylakoid lumen. Plant Cell 23: 304–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schägger H. (2006) Tricine-SDS-PAGE. Nat Protoc 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Schreiber U, Bilger W, Neubauer C. (1994) Chlorophyll fluorescence as a non-intrusive indicator for rapid assessment of in vivo photosynthesis. In E-D Schulze, MM Caldwell, eds, Ecological Studies, Vol 100. Springer-Verlag, Berlin, pp 49–70 [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Töller T, Wolff P, Westhoff P. (2007) The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ozaki Y, Sone N, Feniouk BA, Yoshida M. (2007) The product of uncI gene in F1Fo-ATP synthase operon plays a chaperone-like role to assist c-ring assembly. Proc Natl Acad Sci USA 104: 20776–20781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabayashi A, Ishikawa N, Obayashi T, Ishida S, Obokata J, Endo T, Sato F. (2009) Three novel subunits of Arabidopsis chloroplastic NAD(P)H dehydrogenase identified by bioinformatic and reverse genetic approaches. Plant J 57: 207–219 [DOI] [PubMed] [Google Scholar]
- Takabayashi A, Kadoya R, Kuwano M, Kurihara K, Ito H, Tanaka R, Tanaka A. (2013) Protein co-migration database (PCoM-DB) for Arabidopsis thylakoids and Synechocystis cells. SpringerPlus 2: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima M, Specht M, Hippler M. (2011) The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr Genet 57: 151–168 [DOI] [PubMed] [Google Scholar]
- Thiele A, Winter K, Krause GH. (1997) Low inactivation of D1 protein of photosystem II in young canopy leaves of Anacardium excelsum under high-light stress. J Plant Physiol 151: 286–292 [Google Scholar]
- van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJ. (2004) F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J Cell Biol 165: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmar M, Schlieper D, Winn M, Büchner C, Groth G. (2009) Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J Biol Chem 284: 18228–18235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ballmoos C, Wiedenmann A, Dimroth P. (2009) Essentials for ATP synthesis by F1F0 ATP synthases. Annu Rev Biochem 78: 649–672 [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Gay NJ. (1984) The unc operon: nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta 768: 164–200 [DOI] [PubMed] [Google Scholar]
- Wang ZG, Ackerman SH. (1996) Identification of functional domains in Atp11p: protein required for assembly of the mitochondrial F1-ATPase in yeast. J Biol Chem 271: 4887–4894 [DOI] [PubMed] [Google Scholar]
- Wang ZG, Ackerman SH. (1998) Mutational studies with Atp12p, a protein required for assembly of the mitochondrial F1-ATPase in yeast: identification of domains important for Atp12p function and oligomerization. J Biol Chem 273: 2993–3002 [DOI] [PubMed] [Google Scholar]
- Weber J. (2007) ATP synthase: the structure of the stator stalk. Trends Biochem Sci 32: 53–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. (2007) The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Zeng X, Barros MH, Shulman T, Tzagoloff A. (2008) ATP25, a new nuclear gene of Saccharomyces cerevisiae required for expression and assembly of the Atp9p subunit of mitochondrial ATPase. Mol Biol Cell 19: 1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.