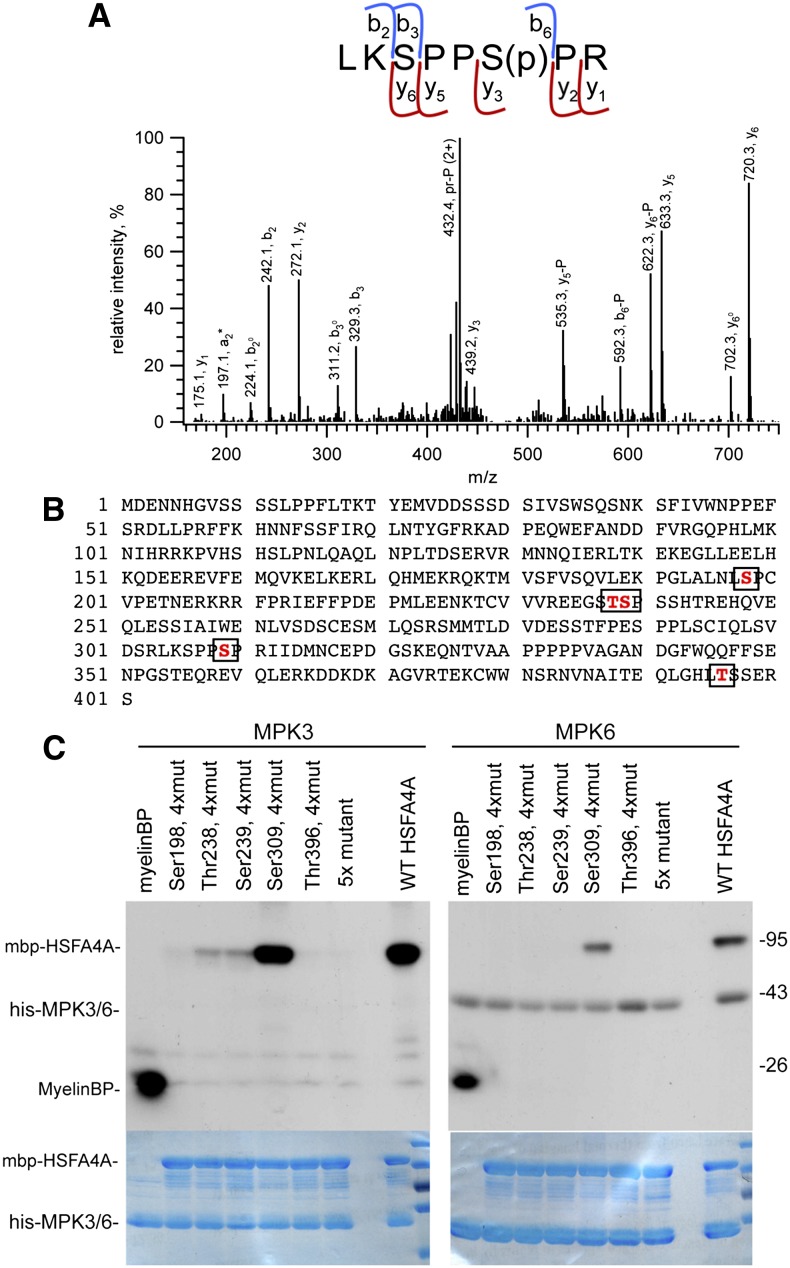
Figure 8.
Identification of HSFA4A phosphorylation sites. A, Identification of HSFA4A phosphorylation site by mass spectrometry. The collision-induced dissociation (CID) spectrum of m/z 481.247 (2+) representing the phosphorylated [304–311] sequence of HSFA4A (Uniprot ID O49403). Fragment ions b3 (unmodified) and y5 and y6 (phosphorylated) unambiguously prove that the site of modification is Ser-6, i.e. Ser-309 of HSFA4A. P denotes the 98-D neutral loss of the phosphate from the corresponding fragment ions. Observed peptide backbone cleavages are indicated in the sequence. B, Amino acid sequence of HSFA4A with MPK3 phosphorylation sites boxed. C, Phosphorylation of wild-type and mutant HSFA4A by MPK3 and MPK6. 5x mutant indicates all identified Ser and Thr residues changed to Ala, and 4xmut indicates all Ser and Thr changed to Ala, except the indicated one. MyelinBP was used in positive control reactions. Top section shows phosphorylation reaction, and bottom section shows Coomassie-stained gels indicating equal loading. [See online article for color version of this figure.]