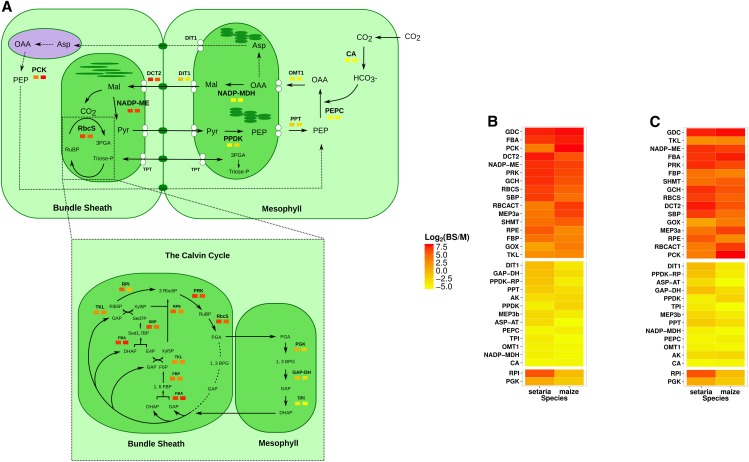
Figure 2.
Abundance of transcripts encoding proteins of the C4 cycle in M and BS cells of S. viridis and maize. A, Summary of transcript quantification in M and BS cells; components of the Calvin-Benson cycle are shown at bottom. Transcripts that are more abundant in the M are colored yellow, while those that are more abundant in the BS are colored red (the scale is shown in the heat map to the right). For each component of the C4 cycle, quantifications for S. viridis and maize are shown on the left and right, respectively. B and C, Log2 fold change of transcript abundance in BS and M cells for all C4 genes sorted by mean enrichment (high to low) in S. viridis and maize (B) or convergence between the two species (C). The top and middle sections represent transcripts that in both species were preferential to BS and M cells, respectively, while the bottom section represents transcripts that showed divergent patterns between the two species. Abbreviations not defined in the text are as follows: AK, adenylate kinase; ASP-AT, Asp aminotransferase; DCT2, dicarboxylate transporter; DIT1, dicarboxylate transporter; FBP, Fru-1,6-bisphosphatase; GCH, Gly cleavage H-protein; GOX, glycolate oxidase; MEP3, putative protein/pyruvate symporter; PPDK-RP, pyruvate,orthophosphate dikinase regulatory protein; PPT, phosphoenolpyruvate/phosphate translocator; RBCACT, Rubisco activase; RbcS, Rubisco small subunit; RPE, ribulose-phosphate3 epimerase; SBP, sedoheptulose-1,7-bisphosphatase; SHMT, Ser hydroxymethyltransferase; TLK, trans-ketolase; TPT, triose phosphate/phosphate antiporter.