Figure. Compare and contrast.
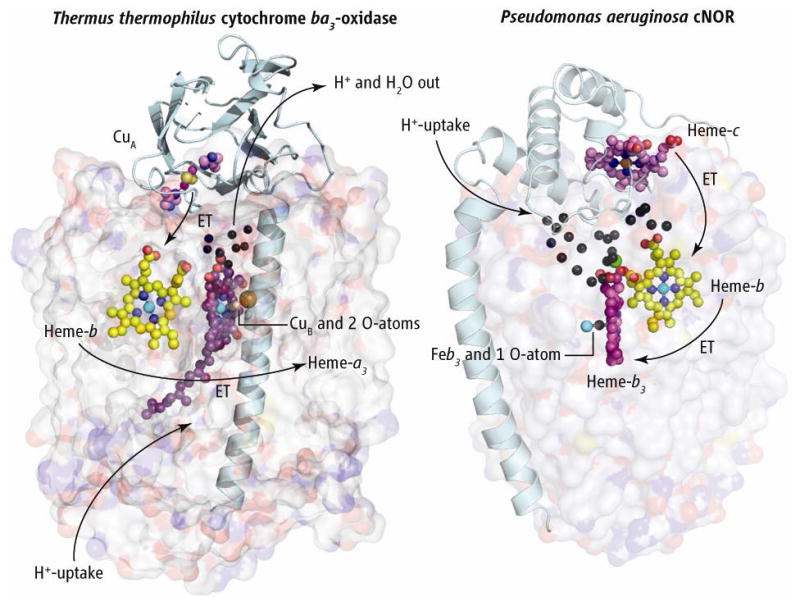
The structures of Thermus thermophilus (Tt) cytochrome ba3 oxidase (left) and cNOR from Pseudomonas aeruginosa (Pa) (right), with large subunits shown as transparent surfaces. The two structures are aligned vertically at the level of the b-hemes and the cNOR structure is rotated approximately 180° about its vertical axis to bring the b-hemes into the same plane. Electrons flow from the subunit II of ba3 that anchors the dinuclear copper cluster CuA, and from the cytochrome c in cNOR. These two redox centers and globular domains exemplify the variability in redox partners in the oxidases and NO reductases, as evolution may allow for occasional interchange of large and small subunits. Electrons flow into the b-hemes before passing to the binuclear active sites. Presumed proton flow in ba3 oxidase and its water exit are represented by arrows. Black spheres represent crystallographically defined water molecules. The single Ca2+ ion is shown as a green sphere. In cNOR, protons appear to flow from the outside into the extensive water cluster, and from there into the binuclear heme-b3/FeB active site.
