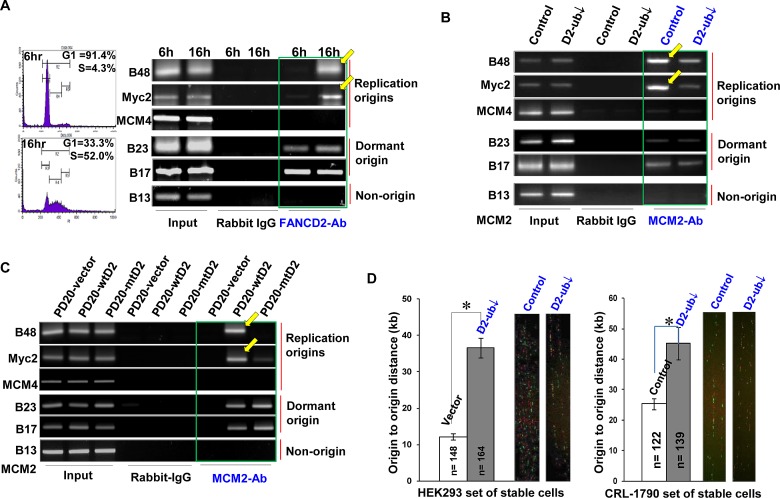
Figure 2. Cells Carrying a Low Basal Level of FANCD2 Monoubiquitination Show a low Amount of Origin DNA associated with MCM2 and a Decreased Rate of Origin-firing.
A. FANCD2 associates with replication origins in an S-phase specific manner. Using FANCD2 antibodies along with rabbit IgG antibody control, we performed ChIP analyses on UV treated as well as starvation-synchronized CRL-1790 cells cultured in regular medium for 6, 12, 14 and 16 hrs, respectively. FANCD2 antibodies can pull down much more B48 and Myc2 origin-containing DNA fragments from the lysates of the pool cells cultured in regular medium for 16 hr and harboring 52% of S-phase cells, as compared to the other pools of cells carrying a lower percentage of S-phase cells. In non-stressed cells, the level of FANCD2 monoubiquitination appears to be positively associated with the amount of pulled-down origin-containing DNA fragments (See FACS data and the basal level of FANCD2 monoubiquitination in Supplementary Fig. 2A1). B. There is a low amount of licensed-origins (origins associated with MCM2) in HEK293 or CRL-1790 cells carrying a reduced basal level of FANCD2 monoubiquitination, as compared to the corresponding control cells carrying a normal basal level of FANCD2 monoubiquitination. Both sets of HEK293 and CRL-1790 stable cells isogenic to FANCL expression (leading to a low basal level of FANCD2 monoubiquitination) were established, which provide two similar systems carrying a normal or reduced basal level of FANCD2 monoubiquitination (Supplementary Fig. 2B1). ChIP assays on these two sets of normally growing CRL-1790 and HEK293 cells were performed. MCM2 antibodies, but not ORC antibodies (Supplementary Fig. 2B2- ORC ChIP assay on CRL-1790 cells; Supplementary Fig. 2B3 – MCM2 and ORC ChIP assays on HEK293 cells), pulled down a low amount of B48 and Myc2-containing DNA fragments in cells carrying a reduced basal level of FANCD2 monoubiquitination, as compared to corresponding control cells containing a normal basal level of FANCD2 monoubiquitination. C. A low amount of origins are associated with MCM2 in FA cells carrying mt or inactivated FANCD2 in comparison with FA cells carrying wtFANCD2. MCM2 ChIP assay on FA derivative cells was performed. These FA derivative cells are PD20 cells (FANCD2-/-) complemented with wtFANCD2, in comparison with mtFANCD2 (K561R); PD220 cells (FANCA-/-, resulting in inactivated FANCD2). MCM2 antibodies only can clearly pull down B48 or Myc2 origin-containing DNA fragments in FA cells carrying wtFANCD2, but not FA cells carrying mt or inactivated FANCD2 (Supplementary Fig. 2B4). D. The origin-firing rate is decreased in normally growing cells carrying a compromised basal level of FANCD2 monoubiquitination. DNA fiber assay was conducted by using CRL-1790 or HEK293 set of stable cells. The distance between fired origins was enhanced in the cells carrying a compromised basal level of FANCD2 monoubiquitination as compared to the corresponding control cells carrying a normal basal level of FANCD2 monoubiquitination. In cells carrying a reduced basal level of FANCD2-monoubiquitination, the number of fired-origins was significantly low as compared to the corresponding control cells (T test, p<0.01). Bars show standard error of the mean (SEM). The images of labeled DNA fibers are shown accordingly at right.