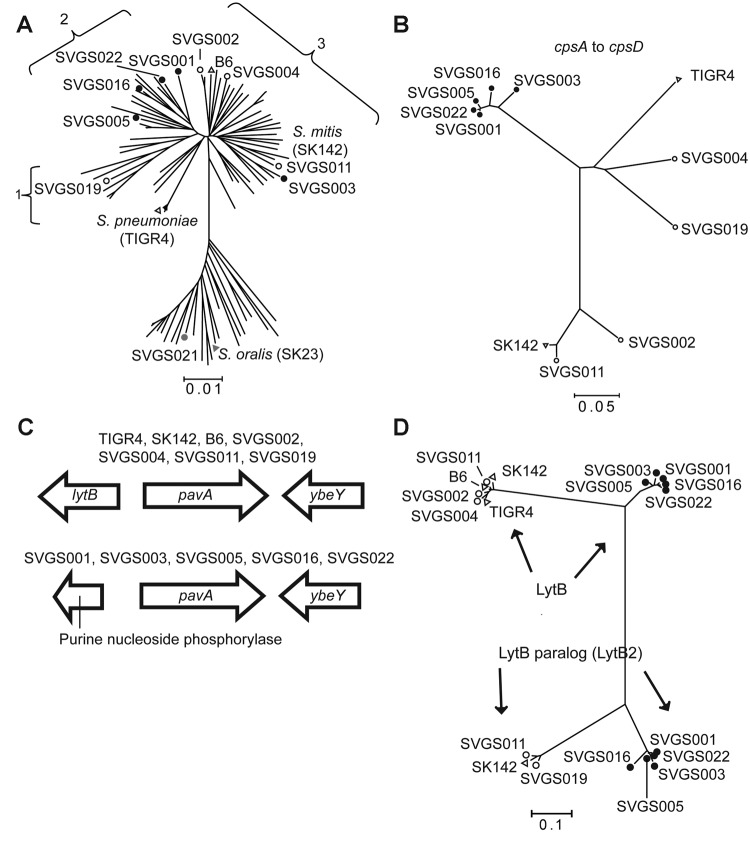
Figure 4.
Selected data from whole-genome analysis of viridans group streptococci (VGS) strains. A) Neighbor-joining tree generated by multilocus sequence analysis (MLSA) of Streptococcus mitis and S. oralis strains, showing locations of VGS strains selected for whole-genome analysis. Numbers 1–3 refer to S. mitis clusters (defined in Figure 3). MLSA locations are also shown for the S. mitis and S. oralis type strains (SK142 and SK23, respectively) and fully sequenced S. mitis strain B6 and S. pneumoniae strain TIGR4. B) SVGS004 mouse challenge data. Neighbor-joining tree of first 4 genes of the capsular polysaccharide encoding operon (cpsA–cpsD). TIGR4 and SK142 are included for reference purposes. Strain B6 is not included because it lacks a cps operon. Note tight clustering of 5 VGS strains (black dots). C) Genetic arrangement surrounding the pavA gene, which encodes a fibronectin-binding protein. Two distinct gene arrangements are present 5′ of the pavA gene, with the arrangement for particular strains as indicated. D) Neighbor-joining tree of LytB protein, which is involved in cell-wall turnover, from fully sequenced S. mitis strains. Some S. mitis strains possess a gene encoding a second LytB-like protein, which we have named LytB2 (ZP_07643922 from strain SK142). Note tight clustering of the same 5 VGS strains (black dots) for the LytB and LytB2 proteins as was observed for the cpsA–cpsD analysis in panel B. A, B, D) SVGS, Shelburne VGS. Scale bars indicate genetic distances.