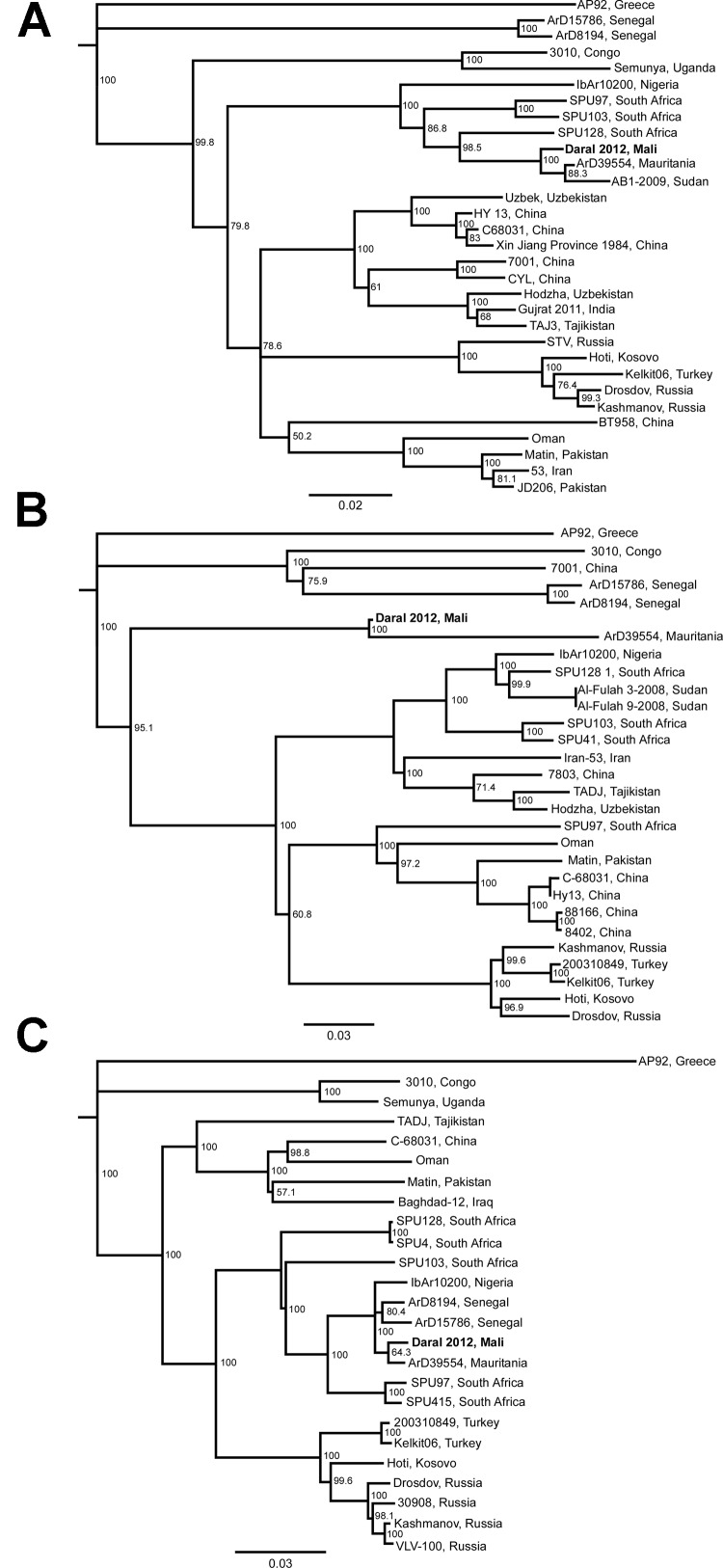
Figure.
Phylogenetic analysis of Crimean–Congo hemorrhagic fever virus (CCHFV) was conducted on the complete nucleoprotein (small genomic segment, nt ≈50–1,500) (A), a 900-bp fragment of the glycoprotein precursor (medium genomic segment, nt ≈4190–5060) (B), and a 1,200-bp fragment of the viral polymerase (large genomic segment, nt ≈590–1760) (C). The fragments were amplified from pooled ticks, and sequence analysis was conducted by using ClustalW (www.ebi.ac.uk/Tools/msa/clustalw2/). Trees were constructed by using the Jukes–Cantor neighbor-joining method with bootstrapping to 10,000 iterations and compared with published sequences of full-length small, medium, and large segments. Bold indicates CCHFV strain from Hyalomma ticks that were collected from cattle at the Daral livestock market near the town of Kati, Mali. Scale bars indicate substitutions per site.