Abstract
Phu Khieo Wildlife Sanctuary (PKWS) is a major hotspot of biological diversity in Thailand but its fungal diversity has not been thouroughly explored. A two-year macrofungal study of this remote locality has resulted in the recognition of a new species of a star-shaped gasteroid fungus in the genus Astraeus. This fungus has been identified based on a morphological approach and the molecular study of five loci (LSU nrDNA, 5.8S nrDNA, RPB1, RPB2 and EF1-a). Multigene phylogenetic analysis of this new species places it basal relative to other Astraeus, providing additional evidence for the SE Asian orgin of the genus. The fungus is named in honour of Her Majesty Princess Sirindhorn on the occasion the 84th birthday of her father, who have both been supportive of natural heritage studies in Thailand.
Introduction
Tropical rain forests are important terrestrial ecosystems. They harbour tremendous biodiversity and several of them are recongized as biodiversity hotspots [1]. Most of the attention paid to this biodiversity has focused on the fauna and flora at the expense of less charasmatic organisms such as fungi. In 1991, Hawksworth [2] has estimated that the number of fungi worldwide ultimately will be around 1.5 million species with fungal diversity considered to be highest in the tropical forests. More recently the estimated number of fungal species has been estimated anywhere between 3.5–5.1 million species [3]. According to Hibbett et al. [4] the overall rate of fungal species discovered worldwide has been fairly level for the last 10 years with a range of 1000–1200 new species reported per year, in both Basidiomycota and Ascomycota but mainly in the latter. Herein a new basidiomycete is added.
The current project is part of an effort to document the diversity of EM fungi associated with a broad range of host plants at a variety of spatial scales in Phu Khieo Wildlife Sanctuary (abbrev.: PKWS) of northeastern Thailand. This project was lead by a team of biologists from Nakhon Phanom University (NPU), in collaboration with Pibulsongkram Rajabhat University (PSRU), Srinakharinwirot University (SWU), Chulalongkorn University (CU), Real Jardín Botánico (RJB-CSIC, Madrid, Spain) and Caledonian Mycological Enterprises (Scotland, UK). The PKWS is a tropical region with a relatively high concentration of ectomycorrhizal associations. It is located in Chaiyaphum province, consisting of a complex of eigth contiguous protected areas in the western part of NE Thailand, and covers and area of 4,594 square kilometers. The western Isan forest complex is the only sizeable expanse of closed forest remaining in the region. It is unique in that it is able to sustain viable populations of wildlife species requiring large home ranges (e.g. tigers and elephants) [5]. It is also important in supporting a range of IUCN Red Listed animals and birds and is essential for conserving water resources in what is otherwise a hot and dry environment [6].
The scant research on fungi for the area has been partly addressed by excursions to describe the macrofungi associated with deciduous and mixed deciduous forest with pine. During the rainy season (July–September) in both 2010 and 2011, a subepigeous, gasteroid fungus was encountered exhibiting characteristics associated with the genus Astraeus Morgan (Order Boletales, clade Sclerodermatineae in [7]) and Geastrum Pers (Order Geastrales in [8]) (Fig. 1). The goal of this study is to identify the phylogenetic placement of this fungus using sequences of LSU nrDNA, 5.8S nrDNA, RPB1, RPB2 and EF1-a, as well as compare its morphology to that of other species of star-shaped gasteroid fungi.
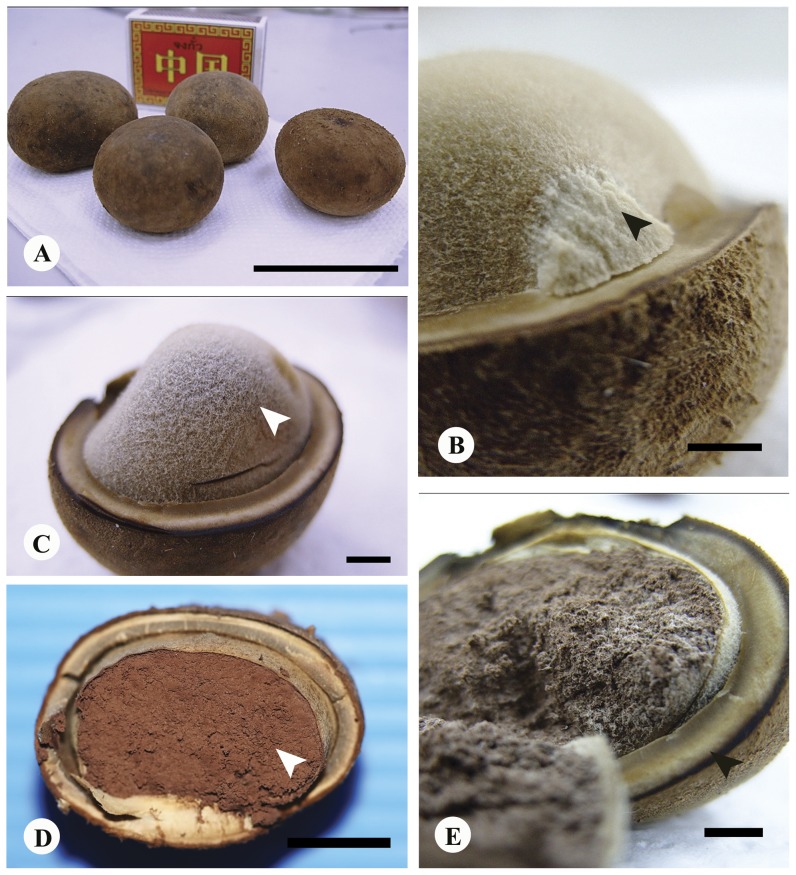
Figure 1. Astraeus sirindhorniae from the field.
(A) immature basidiomes with basal rhizomorphs (arrowhead), bar = 17 mm. (B) mature basidiome split to form a series of rays revealing an endoperidium with an apical opening (arrowhead), bar = 24 mm. (C) basidiospores shooting from an opening apical (in blue circle), bar = 25 mm.
Materials and Methods
Fungal specimens
All necessary permits were obtained for the described field studies issued by Department of National park, wildlife & plant conservation, Bangkok, Thailand (Reference document number 0907.1/17723).
Basidiomes were collected in Phu Khieo Wildlife Sanctuary, Chaiyaphum province, Thailand, during the months of July and September, 2010 and 2011. Field characters such as peridial and glebal colours (Colour identification chart, Royal Botanic Garden, E, 1969) and textures, etc. were recorded in the field and in the laboratory. Basidiospores were mounted in Melzer's reagent [9] and examined and photographed using light microscopy at magnifications of 400–1000× (DIC BX51 Olympus). Mean spore size and range was determined by measuring the diameter of at least 30 spores. Ornamentations were described and later analysed using scanning electron microscopy (SEM). For SEM, spore samples were air-dried, mounted, and sputter-coated with gold before being scanned using a JEOL JSM-840 scanning electron microscope. Peridium structure was examined under polarization microscopy (Imager A1, Zeiss). Attempts to culture the mycelium from fresh basidiomes using a modified Melin Norkrans's medium (MMN) were unsuccessful. Specimens are deposited in BBH, E and MA-Fungi.
DNA isolation, amplification and sequencing
Genomic DNA was extracted from specimens mentioned in Table 1. DNeasy Plant Mini Kit (Qiagen) was used according to the manufacturer's instructions. Five loci were amplified: a) the partial of 5′ end of nuclear ribosomal large subunit RNA gene sequences (nrLSU) with primers LR0R, LR3R, LR5, and LR7 [10]; b) the internal transcribed spacer of nuclear ribosomal DNA (ITS) with primers ITS1F and ITS4B [11]; c) the largest subunit of RNA polymerase II gene sequences (RPB1) with primers RPB1-Af (5′-GAR TGY CCD GGD CAY TTY GG-3′) and RPB1-Cr (5′-CC NGC DAT NTC RTT RTC CAT RTA-3′) [12]; d) the second largest subunit of RNA polymerase II gene sequences (RPB2) with primers RPB2-f5F (5′-GAY GAY MGW GAT CAY TTY GG-3′) [13] and RPB2-b7R (5′-GAY TGR TTR TGR TCR GGG AAV GG-3′) [14]; and e) the transcription elongation factor 1-alpha (EF1-a) with primers 983F (5′-GCY CCY GGH CAY CGT GAY TTY AT-3′) and 2218R (5′-ATG ACA CCR ACR GCR ACR GTY TG-3′) [15]. Polymerase chain reactions (PCR) contained 0.4 U Phire Hot Start II DNA Polymerase (Finnzymes, Sweden), 1× Phire Plant PCR Buffer with 1.5 mM MgCl2, 200 µM of each dNTP and 0.5 µM of each primer. The ITS amplification was run on an Eppendorf thermocycler (Eppendorf, Germany) using the following parameters: initial denaturation of 5 min at 98°C, followed by 40 cycles each with a denaturation step of 5 s at 98°C, annealing for 5 s at 57°C, an elongation step of 20 s at 72°C, and a final elongation step of 10 min at 72°C. The same conditions were used for nrLSU, RPB1, RPB2 and EF1-a amplification except that the annealing temperatures were 50°C, 55°C, 55°C and 57°C, respectively. Amplicons were purified using the QIAquick PCR Purification Kit (Qiagen) and then sequenced at the 1st BASE laboratories Sdn Bhd (Malaysia). Except for RPB2 amplicon was cloned using TA cloning kit (Invitrogen) into Escherichia coli TOP10 before sequenced. Sequences were assembled and edited with BioEdit [16]. BLASTN queries with MEGABLAST option were used to compare sequences obtained against sequences in the National Center of Biotechnology Information (NCBI) nucleotide database [17]. All new sequences have been deposited on the EMBL-EBI database and their accession numbers are presented in Table 1.
Table 1. List of specimens in this study.
| Genus | Species | ID/Herbarium ID | Location/Citation | ITS | nrLSU | RPBI | RPBII | EF1-a |
| Astraeus | sirindhorniae | GAPK1/E30288 | PKWS, Chaiyaphum | HE681772 | HE68182 | HE68191 | KC854536 | KC854542 |
| Astraeus | sirindhorniae | GAPK2/MA-Fungi82080 | PKWS, Chaiyaphum | HE681773 | HE68183 | HE68192 | KC854538 | KC854543 |
| Astraeus | sirindhorniae | GAPK3/BBH34831 | Chaing Mai | HE681774 | HE68184 | HE68193 | KC854539 | KC854544 |
| Astraeus | sirindhorniae | GAPK4/BBH34830 | PKWS, Chaiyaphum | HE681775 | HE68185 | HE68194 | KC854541 | KC854545 |
| Astraeus | asiaticus | Arora 02-121 | Thailand | EU718089 | DQ644199 | FJ536588 | FJ536625 | FJ536665 |
| Astraeus | asiaticus | ASTRAE-44 | Sri Lanka | AJ629395 | ||||
| Astraeus | asiaticus | ASTRAE-56 | Thailand | AJ629396 | ||||
| Astraeus | asiaticus | ASTRAE-64 | Thailand | AJ629400 | ||||
| Astraeus | asiaticus | ASTRAE-65 | Thailand | AJ629401 | ||||
| Astraeus | hygrometricus | Bneil (MB 05-029) | Massachusetts USA | EU718087 | DQ682996 | FJ536586 | FJ536623 | FJ536663 |
| Astraeus | hygrometricus | AWW220a | Massachusetts USA | FJ710187 | ||||
| Astraeus | hygrometricus | ASTRAE-73a | Wisconsin, USA | AJ629398 | ||||
| Astraeus | hygrometricus | ASTRAE-86a | Michigan, USA | AJ629403 | ||||
| Astraeus | hygrometricus | ASTRAE-87b | Greece | AJ629404 | ||||
| Astraeus | hygrometricus | ASTRAE-72b | Spain | AJ629408 | ||||
| Astraeus | hygrometricus | ASTRAE-74a | Wisconsin, USA | AJ629399 | ||||
| Astraeus | hygrometricus | ASTRAE-43 | France | AJ629406 | ||||
| Astraeus | hygrometricus | ASTRAE-42 | France | AJ629394 | ||||
| Astraeus | odoratus | ASTRAE-61 | Thailand | AJ6298776 | ||||
| Astraeus | odoratus | ASTRAE-62 | Thailand | AJ629877 | ||||
| Astraeus | pteridis | Ashy 3 | Switzerland | EU718088 | AF336238 | FJ536587 | FJ536624 | FJ536664 |
| Astraeus | pteridis | PDD88503 | New Zealand | FJ710188 | EU718158 | |||
| Astraeus | pteridis | ASTRAE-36 | Mexico | AJ629392 | ||||
| Astraeus | pteridis | ASTRAE-25 | Wisconsin, USA | AJ629410 | ||||
| Astraeus | pteridis | ASTRAE-24 | Wisconsin, USA | AJ629409 | ||||
| Astraeus | pteridis | ASTRAE-37 | Spain | AJ629393 | ||||
| Boletinellus | merulioides | MB 02-199 | Massachusetts USA | DQ200922 | AY684153 | DQ435803 | DQ366281 | DQ056287 |
| Boletinellus | merulioides | AF336239 | ||||||
| Boletinellus | merulioides | AY612807 | ||||||
| Boletinellus | rompelii | No1192 | EU718159 | |||||
| Calostoma | berkeleyi | AWW268 | Malaysia | EU718090 | EU718128 | FJ536589 | FJ536626 | FJ536666 |
| Calostoma | cinnabarinum | AWW136 | Massachusetts USA | AY854064 | AY645054 | AY780939 | AY857979 | AY879117 |
| Calostoma | Fuscum | OKM 23918 | Western Australia | EU718091 | EU718129 | FJ536590 | FJ536627 | |
| Calostoma | Fuscum | PDD70777 | FJ710190 | EU718161 | ||||
| Calostoma | insignis | Arora 98-31 | Thailand | EU718092 | EU718130 | FJ536628 | ||
| Calostoma | japonicum | TKG-SC-40701 | Japan | EU718093 | EU718131 | FJ536591 | FJ536629 | |
| Calostoma | junghuhnii | VC1151 | India | EU718163 | ||||
| Calostoma | lutescens | 1329 | FJ710192 | EU718164 | ||||
| Calostoma | orirubra | HKAS32119 | China | FJ710195 | EU718165 | |||
| Calostoma | rodwayi | GMM 7572 | New Zealand | EU718095 | EU718133 | FJ536631 | ||
| Calostoma | sarasinii | DED7660 | Malaysia | EU718096 | EU718134 | FJ536593 | FJ536632 | FJ536668 |
| Calostoma | Sp | HKAS38133 | China | EU718097 | EU718135 | FJ536633 | ||
| Calostoma | Sp | HKAS38139 | China | EU718098 | EU718136 | FJ536594 | FJ536634 | |
| Diplocystis | wrightii | DH2002 | DQ534665 | |||||
| Gyroporus | aff. castaneus | E4600 | EU718169 | |||||
| Gyroporus | aff. castaneus | E843c | EU718170 | |||||
| Gyroporus | aff. castaneus | E4879c | FJ710208 | |||||
| Gyroporus | castaneus | Gc1 | Germany | EU718099 | AF336252 | FJ536595 | FJ536635 | FJ536669 |
| Gyroporus | castaneus | 239-97 | USA | EU718100 | AF336253 | FJ536596 | FJ536636 | FJ536670 |
| Gyroporus | castaneus | REH8804 | Thailand | EU718101 | EU718137 | FJ536597 | FJ536637 | FJ536671 |
| Gyroporus | aff. cyanescens | REH8821 | Western Australia | EU718103 | EU718139 | FJ536599 | FJ536639 | FJ536673 |
| Gyroporus | aff. cyanescens | E486 | Australia | EU718173 | ||||
| Gyroporus | cyanescens | MB 05-001 | USA | EU718102 | EU718138 | FJ536598 | FJ536638 | FJ536672 |
| Gyroporus | cyanescens | Gcy2 | Germany | AF336254 | ||||
| Gyroporus | cyanescens | E8758c | Australia | EU718171 | ||||
| Gyroporus | aff. cyanescens | OKM23719 | Western Australia | EU718104 | EU718140 | FJ536600 | FJ536640 | |
| Gyroporus | purpurinus | PRL 3737 | Illinois, USA | EU718105 | EU718141 | FJ536601 | FJ536641 | FJ536674 |
| Gyroporus | sp. | REH8799 | Thailand | EU718106 | EU718142 | FJ536602 | FJ536642 | FJ536675 |
| Gyroporus | sp. | Arora 00-429 | Zimbabwe | EU718107 | EU718143 | FJ536603 | FJ536643 | FJ536676 |
| Gyroporus | sp. | E8155 | EF561627 | |||||
| Gyroporus | sp. | REH8805 | EU718175 | |||||
| Gyroporus | subalbellus | OKM25477 | Texas, USA | EU718108 | EU718144 | FJ536604 | FJ536644 | FJ536677 |
| Phlebopus | beniensis | Omon 98.015 | AY612822 | |||||
| Phlebopus | marginatus | REH8883 | Eastern Australia | EU718109 | EU718145 | FJ536605 | FJ536645 | FJ536678 |
| Phlebopus | marginatus | MEL2145841 | Australia | FJ600322 | ||||
| Phlebopus | portentosus | php1 | Africa | EU718110 | AF336260 | FJ536606 | FJ536646 | FJ536679 |
| Phlebopus | sp. | AY612816 | ||||||
| Phlebopus | sp. | REH8795 | Thailand | EU718111 | AF336260 | FJ536607 | FJ536647 | FJ536680 |
| Phlebopus | sudanicus | AF336261 | ||||||
| Pisolithus | albus | PERTH4681 | Australia | FJ710202 | EU718176 | |||
| Pisolithus | arhizus | AF336262 | ||||||
| Pisolithus | aurantioscabrosus | AWW297 | Malaysia | EU718112 | EU718146 | FJ536608 | FJ536648 | FJ536681 |
| Pisolithus | sp. | ECV3205 | California USA | EU718113 | EU718147 | FJ536609 | FJ536649 | |
| Pisolithus | tinctorius | AWW219 | Massachusetts USA | EU718114 | EU718148 | FJ536610 | FJ536650 | FJ536682 |
| Scleroderma | areolatum | AWW211 | Massachusetts USA | EU718115 | EU718149 | FJ536611 | FJ536651 | FJ536683 |
| Scleroderma | areolatum | PBM2208 | W. Australia | EU718116 | EU718150 | FJ536612 | FJ536652 | FJ536684 |
| Scleroderma | bermudense | BZ3961 | Belize | EU718118 | DQ644137 | FJ536614 | FJ536654 | FJ536686 |
| Scleroderma | citrinum | AWW212 | Massachusetts USA | EU718119 | EU718151 | FJ536615 | FJ536655 | FJ536687 |
| Scleroderma | citrinum | AF336266 | ||||||
| Scleroderma | columnare | AF261533 | ||||||
| Scleroderma | columnare | AF336273 | ||||||
| Scleroderma | echinatum | AF336268 | ||||||
| Scleroderma | fuscum | Trappe26575 | EU718178 | |||||
| Scleroderma | leave | MCA242 | North Carolina USA | EU718117 | DQ677138 | FJ536613 | FJ536653 | FJ536685 |
| Scleroderma | leave | OSC27936 | EU718120 | DQ683003 | FJ536616 | |||
| Scleroderma | mcalpinei | OSC 24605 | EU718122 | DQ682999 | FJ536657 | |||
| Scleroderma | meridionale | AWW218 | Massachusetts USA | EU718121 | EU718152 | FJ536617 | FJ536656 | FJ536688 |
| Scleroderma | polyrhizum | AWW216 | Massachusetts USA | EU718123 | EU718153 | FJ536618 | FJ536658 | FJ536689 |
| Scleroderma | sinnamariense | AWW254 | Malaysia | EU718124 | EU718154 | FJ536619 | FJ536659 | FJ536690 |
| Scleroderma | sp. | HKAS43607 | FJ710210 | |||||
| Scleroderma | sp. | Arora9917 | EU718179 | |||||
| Scleroderma | sp. | MCA2168 | EU718180 | |||||
| Scleroderma | sp. | MEL2295738 | EU718181 | |||||
| Scleroderma | sp. Brown | AWW311 | Malaysia | EU718126 | EU718156 | FJ536621 | FJ536661 | FJ536692 |
| Scleroderma | sp. White | AWW260 | Malaysia | EU718125 | EU718155 | FJ536620 | FJ536660 | FJ536691 |
| Scleroderma | verrucosum | AF336271 | ||||||
| Tremellogaster | surinamensis | MCA 1985 | Guyana | EU718127 | DQ534664 | FJ536622 | FJ536662 | FJ536693 |
Phylogenetic analysis
Two datasets were created for this study. One is a multigene dataset that examines the phylogenetic position of the gasteroid fungus from PKWS using ribosomal RNA and protein coding genes (nrLSU, the 5.8S region of the ITS, RPB1, RPB2 and EF1-a). Genes missing for individual samples were coded as “?” in the dataset to represent missing data. A second dataset consisted of only ITS sequence data to compare this taxon against other known Astraeus species. Both datasets, consisting of original sequences, plus sequences acquired from Genbank, were aligned using MUSCLE [18] with additional manual adjustments to the alignment performed in Mesquite 2.74 [19].
For each dataset, maximum likelihood and Bayesian analyses were performed using the CIPRES web portal (http://www.phylo.org/portal2/) [20]. Maximum likelihood bootstrapping analyses was performed on each dataset with RAxML 7.2.8 [21], using the default parameters as implemented on the CIPRES NSF XSEDE resource with bootstrap statistics calculated from 1000 bootstrap replicates. Bayesian phylogenetic analyses were performed using Mr Bayes v. 3.2.1 [22] on CIPRES XSEDE resource with default parameters (Nst = 6, with 2 runs, 4 chains per run, each run searching for 1000000 generations sampling every 1000th generation).
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS ONE article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
The new taxon described herein has been submitted to MycoBank and the unique MycoBank number provided can be used to retrieve the associated taxonomic information at http://www.mycobank.org/MycoTaxo.aspx?Link=T&Rec=.
Results
Phylogenetic analysis
BLAST searches with megablast option were used to compare the sequences obtained (nrLSU and RPB1 around 1460 and 1310 bp, respectively) against the sequences in the National Center of Biotechnology Information (NCBI) nucleotide databases [17]. Sequences from the gasteroid fungus from PKWS produce matches for Astraeus spp., Diplocystis wrightii Berk. & M.A. Curtis, Pisolithus spp., Scleroderma spp., Tremellogaster surinamensis E. Fisch. and Veligaster columnaris (Berk. & Broome) Guzman. All of these taxa are gasteroid Boletales included in Sclerodermatineae [7].
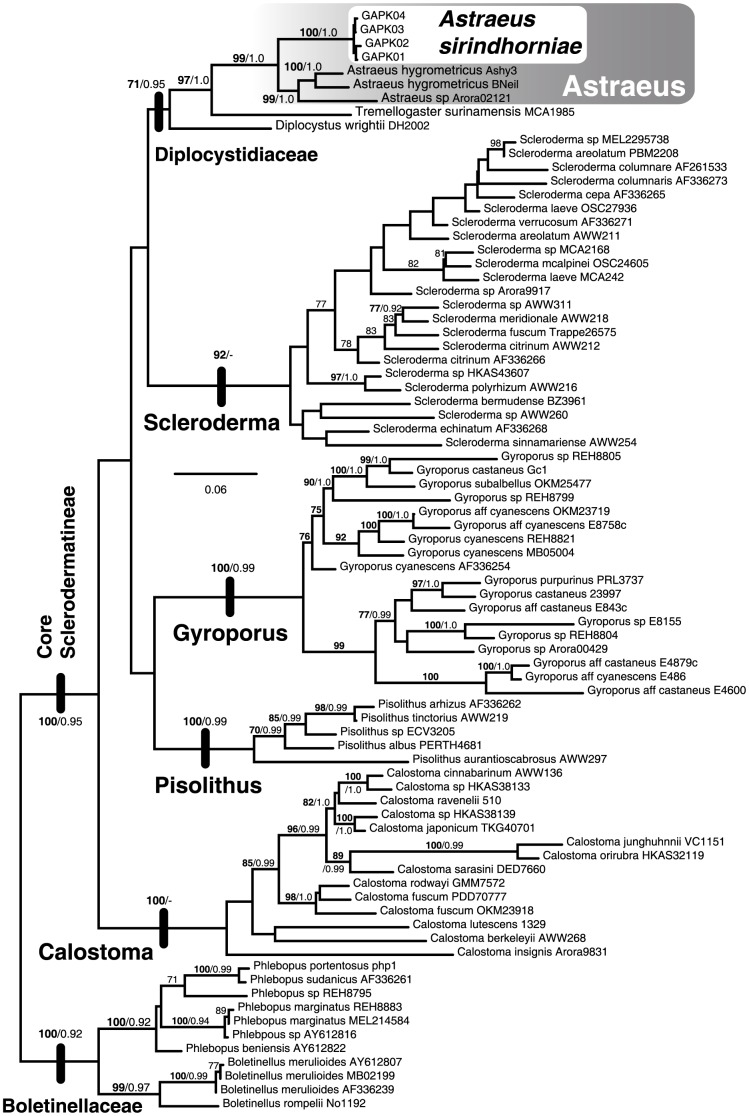
To evaluate the phylogenetic position of the sclerodermatoid fungus from PKWS, a multigene dataset was created using nrLSU, 5.8S, RPB1, RPB2 and EF1-a genes from 80 specimens. This dataset was rooted using the Boletinellaceae (Boletellus and Phlebopus) while the genera Astraeus, Calostoma, Diplocystis, Gyroporus, Phlebopus, Scleroderma and Tremellogaster consisted of the ingroup. Maximum likelihood bootstrap (MLB) and Bayesian posterior probabilities (PP) strongly support a monophyletic placement for the sclerodermatoid fungus with Astraeus (MLB = 99%, PP = 1.0; Fig. 2). With it's inclusion in Astraeus, there is a strong sister relationship with the monotypic genus Tremellogaster (MLB = 97%, PP = 1.0) and weak support for the inclusion of these taxa, along with Diplocystus to form the Diplocystidiaceae (MLB = 71%, PP = 0.95). Sequences use for both phylogenetic datasets and their corresponding GenBank accession numbers are given in Table 1.
Figure 2. Maximum likelihood tree from a multigene dataset reveals the placement of Astraeus sirindhorniae within the Sclerodermatineae.
Thick vertical black bars identify root branch for the taxonomic lineage indicated by the adjacent label. Numbers above branches identify the statistics bootstrap percentages (bold text, before forward slash) and Bayesian posterior probabilities (normal text, after forward slash) for that branch. Maximum likelihood bootstraps from 1000 iterations. Bayesian posterior probabilities from 1000 iterations (1 million runs sampling every 1000th iteration).
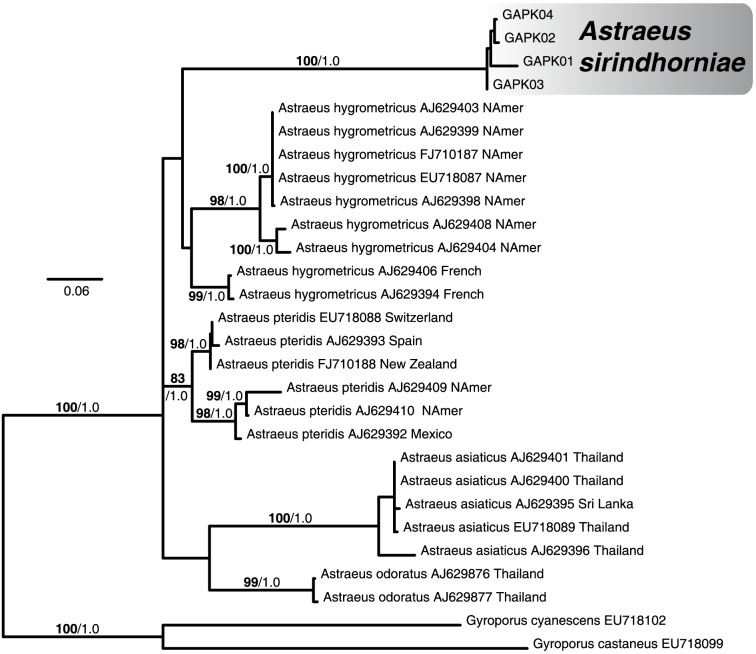
An ITS dataset was developed to evaluate the uniqueness of this new taxon relative to other Astraeus species. This dataset consists of 28 samples (2 outgroup samples from Gyroporus). Maximum likelihood and Bayesian phylogenetic analysis identifies eight major clades that can be recognized as species (each with MLB>98% and PP = 1.0; Fig. 3). Five of these represent taxa already defined by Phosri et al. [23]. Four samples of the new Astraeus taxon form a strongly supported group distinct from the other major Astraeus clades (MLB = 100%, PP = 1.0) which we will from now on refer to as Astraeus sirindhorniae.
Figure 3. Maximum likelihood tree from ITS dataset identifies Astraeus sirindhorniae as a distinct species of Astraeus.
Numbers above branches identify the statistics bootstrap percentages (bold text, before forward slash) and Bayesian posterior probabilities (normal text, after forward slash) for that branch. Maximum likelihood bootstraps from 1000 iterations. Bayesian posterior probabilities from 1000 iterations (1 million runs sampling every 1000th iteration).
Taxonomy
Astraeus sirindhorniae sp. nov. Watling, Phosri, Sihanonth, A.W.Wilson & M.P. Martín
Mycobank
MB803956
Etymology
The species is named after Princess Sirindhorn on the occasion the 84th birthday of her father, who have both been supportive of natural heritage studies in Thailand and as a token of respect and recognition of the great interest shown by Her Majesty in the natural history and conservation of natural resources of Thailand. Now her name will be known in association with the Greek Titan of Astrology (Astraeus).
Holotype
Thailand, Phu Khieo Wildlife Sanctuary, Chaiyaphum, coll. C.Phosri, 9 September 2010, (BBH34830)
Diagnostic description
Basidiomycota: Boletales: Sclerodermatineae
Large, subglobose to ellipsoid, subepigeous, dry basidiomes splitting at maturity to form a non-gelatinised, exoperidium with rays that unfold into a star-shaped structure. Enclosed within the exoperidium is a pale, thin, dry, stipitate endoperidium containing a powdery gleba of date-brown to umber (Colour identification chart, Royal Botanic Garden, E, 1969), large, globose, distinctly but minutely verrucose spores <11 µm diam. and lacking a columella.
Basidiomes subglobose to ellipsoid at first (Fig. 4A), slightly compressed, hard, subepigeous 24.5–55.0 mm diam., dry, with woolly, adpressed covering forming felty, adpressed triangular to hexagonal scales, denser and more fluffy towards base where they are intermixed with date-brown to sepia rhizomorphs (Fig. 1A), splitting into concentric zones which fuse towards uppermost, exoposed parts, with thick, complex exoperidium (Fig. 4E), expanding to become star-shaped and then 40–100 mm broad, tough, surface often encrusted with soil particles; odour strong, penetrating, pleasant. When mature exoperidium buff to snuff-brown, squamulose, consisting of at least 3 distinct layers 3–5 mm thick when fresh, contracting to <1 mm when dry, leathery, splitting into 6–8 broad, stellate rays, innermost layer varying from buff to brownish, extensively scaly cracked to give almost regular pattern. Endoperidium shortly stipitate (Fig. 4B), globose to subglobose ca 18–32 µm diam., white at first, becoming buff to hazel when mature, very fluffy-fibrillose (Fig. 4C), even velvety opening by apical, irregular tear and lacking defined peristome. Gleba purplish chestnut becoming umber to date-brown when mature (Fig. 4D), lacking columella. Exoperidial suprapellis ca 70–80 µm, brownish, consisting of interwoven, periclinal to perpendicular, thin- or thick-walled hyphae 4–7 µm broad, with central lumen and walls 1–2 µm thick (Fig. 5A–C). Exoperidial mediopellis, fibrous ca 600 µm broad of interwoven periclinal to orthogonal hyphae 5–7 µm broad with hyaline, continuous lumen and walls 1–2 µm thick and becoming more parallel at junction with subpellis (Fig. 5C–F). Exoperidial subpellis pseudoparachymatous, ca 1050–1100 µm broad of hyaline, parallel to anticlinal, thick-walled hyphae with walls 1–2 µm thick (Fig. 5F and Fig. 6A–B). Clamp-connections absent in exoperidium. Endoperidium consisting of brownish, interwoven, unbranched, aseptate hyphae ca 5–8 µm broad with continous lumen and walls 1–2 µm thick, clamp-connections absent. Capillitium of long, unbranched, interwoven, hyaline, aseptate threads 4–7 µm broad and lacking clamp-connections (Fig. 6D). Basidiospores globose, (5.19)-6–11 µm diam., including ornamentation, umber to date-brown (Fig. 6D), with moderately dense, rounded, narrow, tapered, separate tubercles which coalesce in groups (Fig. 6 E–F).
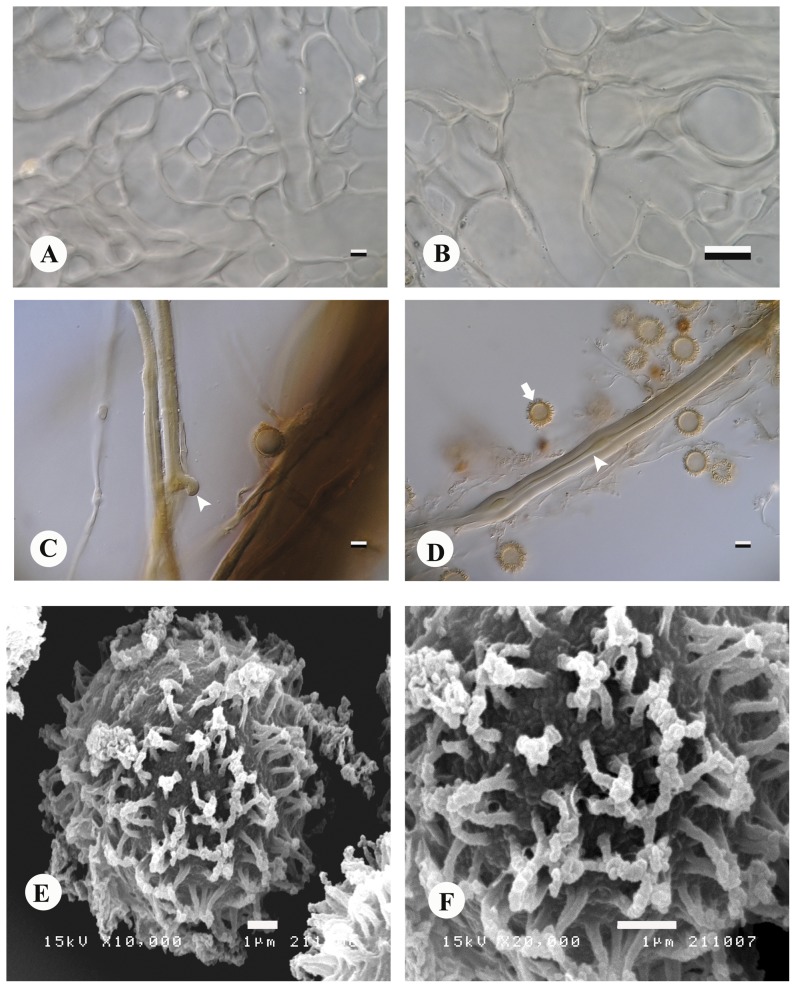
Figure 4. Astraeus sirindhorniae.
(A) immature basidiomes, bar = 60 mm. (B) short stipitate endoperidium (arrowhead), bar = 3 mm. (C) fibrillose endoperidium (arrowhead), bar = 3 mm. (D) gleba colour become umber to date- brown when mature (arrowhead), bar = 10 mm. (E) complex outer peridium, bar = 3 mm.
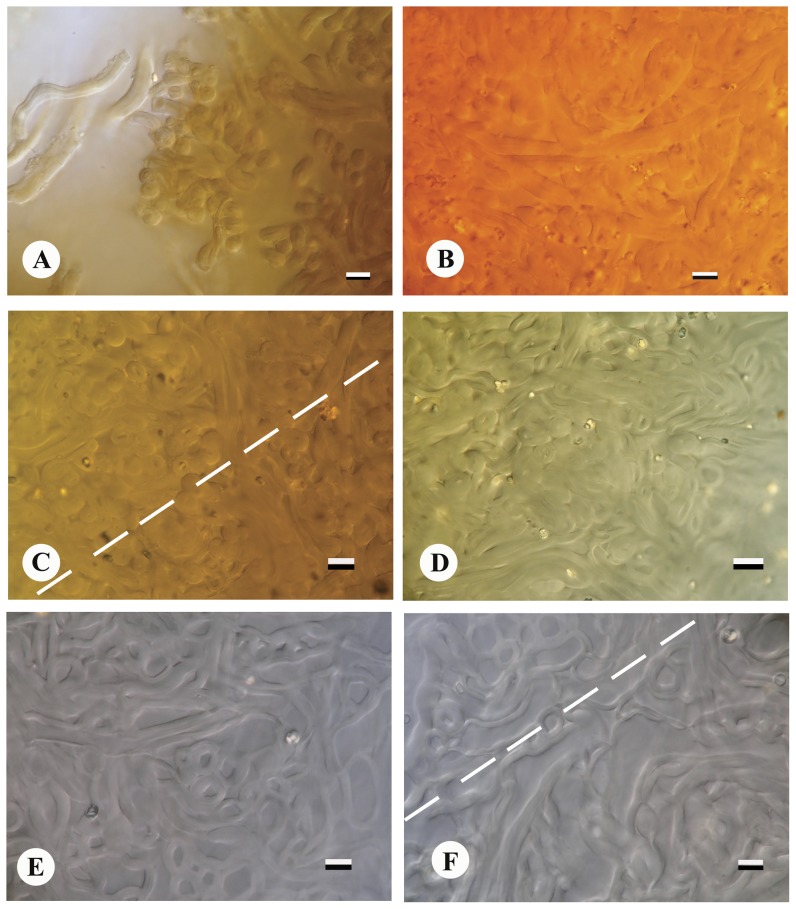
Figure 5. Astraeus sirindhorniae.
Exoperidium layers. (A) exoperidial suprapellis, outer most surface, bar = 6 µm. (B) exoperidial suprapellis, bar = 7 µm. (C) interface layer between exoperidial suprapellis (top left) and mediopellis (lower right), bar = 6 µm. (D) exoperidial mediopellis, bar = 7 µm. (E) exoperidial mediopellis (inner most), bar = 7 µm. and (F) interface layers between exoperidial mediopellis (top left) and subpellis (lower right), bar = 8 µm. Magnification at 1,000×.
Figure 6. Astraeus sirindhorniae.
Exoperidium layers (A–B). (A) exoperidial subpellis, bar = 5 µm. (B) exoperidial subpellis (innermost), bar = 10 µm. (C) rhizomorph hyphae with clamp connection (arrowhead), bar = 5 µm. (D) capillitium hyphae displaying continuous lumen (arrowhead) and basidiospore (arrow), bar = 5 µm. (E–F) spore ornamentation demonstrated coalescent spines in groups, bar = 1 µm. A–D magnification at 1,000×.
Habitat
In rainy season, gregarious, partially buried in ultisols in dry deciduous forests associated with Dipterocarpus tuberculatus Roxb., Shorea obtusa Wall. and Shorea siamensis Miq.
Distribution
North and Northeastern areas of Thailand.
Material examined
Thailand, Chiyaphum province, Phu khieo Wildlife Sanctuary, Dipterocarp forests, N 16°27′32″ and E 101°39′414″, elev. 640 msl, 9 September 2010 (BBH 34830, duplicate E30288, duplicate MA-Fungi 82080); Ibidem, date, (BBH 34831), Mae Cham district, Dipterocarp forests, N 18°31′981″ and E 98°24′939″, June–September 2010.
Note
Her Royal Highness the Crown Princess of Thailand, has considered and granted for a new fungus name; A. sirindhornii. This name is a great honor and a privilege. However according to ICBN Recommendation 60C.1(b) If the personal name ends with a consonant (but not in -er), substantival epithets are formed by adding -i- (stem augmentation) plus the genitive inflection appropriate to the sex and number of the person(s) honoured (e.g. lecard-ii for Lecard (m), wilson-iae for Wilson (f), verlot-iorum for the Verlot brothers, braun-iarum for the Braun sisters, mason-iorum for Mason, father and daughter). Therefore A. sirindhornii should ending with –iae and then the epithet to be spelled; A. sirindhorniae.
Discussion
Astraeus sirindhorniae represents a new species of star-shaped gasteroid fungus which differs morphologically from many other genera of star-shaped fungi. In comparing this species, the earthstar genus, Geastrum, tends to have a well defined peristome. Myriostoma species may be distinguished by the formation of multiple irregular shaped peristomes from which spores escape. Trichaster differs in having an endoperidium that remains attached to the exoperidium after opening, then soon disintegrates leaving a powdery spore-mass suppported by a stout, persistent collumella. The endoperidium of Terrostella is thin and peels away to expose a powdery spore-mass supported by a distinct sterile base. Phialastrum has a strongly developed columella and Geasteropsis produces an extremely hard basidiome when dry.
According to Phosri et al. there are only two Astraeus species in Thailand, A. odoratus and A. asiaticus [23], [24]. Astraeus odoratus is found under ecological conditions similar to those at the Phu Khieo Wildlife Sanctuary. However, A. sirindhorniae differs in its much larger basidiomes, both when immature and when its rays are fully expanded, displaying flared margins, and exposing complex layering. Astraeus sirindhorniae is further differentiated from A. odoratus through the presence of prominent rhizomorphs, a complex multi-layered exoperidium, and smaller basidiospores (range 6–11 µm). These basidiospores are also smaller than A. asiaticus spores (8.75–15.2 µm) and generally given for A. hygrometricus s. str. viz. (7.5–12 µm) [25], [26], [27], [28], [29]. The spore ornamentation of A. sirindhorniae is notable under SEM as it has moderately dense, rounded, narrow, tapered, separate tubercles, which coalesce spines in groups. In addition, A. sirindhorniae has a short stipitate, very fluffly- fibrillose endoperidium when immature, which further differentiates this taxon from other Astraeus species.
The outermost felty, scaly covering of the young basidiomes of A. sirindhorniae closely resembles that of members of Scleroderma previously placed in Veligaster. The gleba is probably not divided into tramal plates. As in A. sirindhorniae clamp-connections are absent from both the gleba and the peridium. On maturing the highly gelatinized middle layer is exposed well before the powdery gleba is revealed. In A. sirindhorniae the peridial medio- and subpeillis are not gelatinized and the hyphae are fully differentiated but otherwise the very young basidiomes are similar in primordial structure.
In the multi-gene phylogenetic analyses A. sirindhorniae, along with other Astraeus species, form a monophyletic clade with Tremellogaster, and Diplocystis (Fig. 2). This clade is recognized as the Diplocystidiaceae. The structure of the peridium in Tremellogaster is also rather complex: the outer wall consists of thickened, sclerotised hyphae; the middle layer is brown and heavily gelatinised and divided into polygonal areas of plate-like, non-gelatinous tissue; and the innermost layer consisting of hyaline, thin-walled hyphae similar to those in A. sirindhorniae but posses transverse thickenings. A summary of the pertinent characters and literature references for Tremellogaster are given in Watling [30].
Members of the Sclerodermatineae form ectomycorrhizal associations with many host plants. Species of Astraeus are known to associate with ectomycorrhizal plant hosts in the Pinaceae, Betulaceae, Fagaceae, Ericaceae and Dipterocarpaceae [31]. Given its phylogenetic placement, and the fact that it is found in dipterocarp dominated forests, it is likely that A. sirindhorniae is also an ectomycorrhizal fungus. Further study into the ecology of this species is needed in order to conclusively identify possible relationships to dipterocarpacious hosts.
In the multigene phylogeny the basal position of A. sirindhorniae relative to other Astraeus taxa is interesting from a biogeographic standpoint (Fig. 2). This placement suggests a Southeast Asian origin for the genus, which is observed in many Sclerodermatineae genera [31]. However, this is complicated by the fact that the basal Diplocystidiaceae (Diplocystis and Tremellogaster) are monotypic genera whose species are described from the new world (the Caribbean and South America respectively). Further investigation into the biogeogaphic history of these taxa is necessary to understand the current distribution of new- and old-world Astraeus.
Conclusions
In summary A. sirindhorniae is morphologically distinguished from A. odoratus, A. asiaticus and A. hygrometricus s.l. by basidiome and basidiospore size, spore ornamentation and peridium structure. Phylogenetic analysis clearly resolves Astraeus sirindhorniae as a basal lineage of Astraeus, within the Diplocystidiaceae and Sclerodermatineae. This systematic relationship, in combination with its associations with dipterocarp forests, it is probable that this species is ectomycorrhizal with members of the Dipterocarpaceae. Astraeus sirindhorniae represents a new gasteroid, star-shaped fungus from Thailand. This discovery reinforces the belief that fungi represent a group of organisms with many undescribed taxa; some of which exist within the dry evergreen dipterocarp forests of SE Asia.
Acknowledgments
First and foremost, we would like to thank Her Royal Highness the Crown Princess of Thailand, Princess Sirindhorn for her considered and permission for a new fungus name. This name is a great honor and a privilege. We thank to a former Head of Phu Khieo Wildlife Sanctuary, Dr. Kanjana Nitaya, their staffs and Dr. Rungpetch Kaengraeng, Nittaya Tunpin, Suchart Junthahum, Aor Jorn-em, Apichai Nisaiparm, Preeyaporn Dokmai for facilitated the excursions. We are grateful to Asistant Professor Jittra Piapukieow and Miss Somsri Rinjai who supported us the materials for study. We thank Associated Professor Prakitsin Sihanonth for providing SEM photographs, Catherine Amie and Tharnrat Kaewkrajang for Tremelogaster surinamensis and Astraeus odoratus photographs. C. Phosri wishes to thank Associate Professor Akiyoshi Yamada, Laboratory of Mycorrhizas, Shinshu University, for allowing him the microscopic study with sophisticated equipments during his 3 months stay in Japan. C. Phosri would also like to thank Kongsak Deethongtong and Joss Friedrich Kurz for their assistance with photography and artwork. C. Phosri and M.P.Martín thank Biod-Iberia and Synthesys I projects and the Masumae International Foundation (MIF) for the opportunities in establishing international cooperations. We are grateful to the anonymous reviewers for their helpful comments in reviewing this manuscript.
Funding Statement
This study was funded in part by National Research Council of Thailand (NRCT) and the Masumae International Foundation (MIF) to C. Phosri. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.ASEAN Centre for Biodiversity (2010) ASEAN biodiversity outlook. Dolmar Press, Inc., Philippines, 208 pp. [Google Scholar]
- 2. Hawksworth DL (1991) The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol Res 95: 641–655. [Google Scholar]
- 3. O'Brian H, Parrent JL, Jackson J, Moncalvo J-M, Vilgalys R (2005) Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol 71 (9) 5544–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hibbett DS, Ohman A, Kirk PM (2009) Fungal ecology catch fire. New Phytol 184: 279–282. [DOI] [PubMed] [Google Scholar]
- 5.RFD & Kasetsart (2002) Socioeconomic and landuse surveying of Phukhieo wildlife sanctuary and buffer zone by using remote sensing and GIS. Kasetsart University, Royal Forest Department, Bang Khen. [Google Scholar]
- 6.Mongkol K, Kitti K, Wonchanok S, Nipon S (1999) Biodiversity of vertebrate animals in Phu Khieo wildlifes. Phu khieo Wildlife Sanctuary, Natural resource Conservation office, Royal Forest Department, Chaiyapum. [Google Scholar]
- 7. Binder M, Hibbett DS (2006) Molecular systematics and biological diversification of Boletales. Mycologia 98: 971–981. [DOI] [PubMed] [Google Scholar]
- 8. Hosaka K, Bates ST, Beever R, Castellano MA, Colgan W III, et al. (2006) Molecular phylogenetics of the gomphoid-phalloid fungi with an establishment of the new subclass Phallomycetidae and two new orders. Mycologia 98 (6) 949–959. [DOI] [PubMed] [Google Scholar]
- 9.Largent D, Johnson D, Watling R (1977) How to identify mushrooms to genus III. Microscopic features. Mad River Press, Eureka, California, USA. [Google Scholar]
- 10. Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gardes M, Bruns TD (1993) ITS primers with enhanced specify for Basidiomycetes: application to identification of mycorrhizae and rusts. Mol Ecol 2: 113–118. [DOI] [PubMed] [Google Scholar]
- 12. Matheny PB, Liu YJ, Ammirati JF, Hall BD (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot 89: 688–698. [DOI] [PubMed] [Google Scholar]
- 13. Liu YL, Whelen S, Hall BD (1999) Phylogenetic relationships amomg ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- 14. Matheny PB (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol Phylogenet Evol 35: 1–20. [DOI] [PubMed] [Google Scholar]
- 15. Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- 16. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- 17. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar R (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maddison WP, Maddison DR (2008) Mesquite: a modular system for evolutionary analysis. Version 2.74. Available: http://mesquiteproject.org.
- 20.Miller MA, Holder MT, Vos R, Midford PE, Liebowitz T, et al. (2009) The CIPRES Portals.
- 21. Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 (21) 2688–2690. [DOI] [PubMed] [Google Scholar]
- 22. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61 (3) 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Phosri C, Martín MP, Sihanonth P, Whalley AJS, Watling R (2007) Molecular study of the genus Astraeus . Mycol Res 111: 275–286. [DOI] [PubMed] [Google Scholar]
- 24. Phosri C, Watling R, Martín MP, Whalley AJS (2004) The genus Astraeus in Thailand. Mycotaxon 89 (2) 453–463. [Google Scholar]
- 25.Coker WC, Couch JN (1928) The gasteromycetes of the Eastern United States and Canada. University of North Carolina Press, Chapel Hill. 201 p. [Google Scholar]
- 26.Cunningham GH (1944) Gasteromycetes of Australia and New Zealand, Dunedin. 236 p. [Google Scholar]
- 27.Liu B (1984) The gasteromycetes of China. Vaduz, Germany, Nova Hedwigia. [Google Scholar]
- 28.Lloyd CG (1902) The Geastrae. Cincinnati, 43 p. [Google Scholar]
- 29. Nouhra ER, De Toledo LD (1998) The first record of Astraeus hygrometricus from Argentina. Mycologist 12 (3) 112–113. [Google Scholar]
- 30. Watling R (2008) A manual and source book on the Boletales and their allies. Synopsis Fungorum 24: 1–245. [Google Scholar]
- 31. Wilson AW, Binder M, Hibbett DS (2012) Diversity and evolution of ectomycorrhizal host associations in the Sclerodermatineae (Boletales, Basidiomycota). New Phytol 194 (4) 1079–1095. [DOI] [PubMed] [Google Scholar]
- 32. Phosri C, Martín MP, Watling R (2013) Astraeus: hidden dimensions. IMA Fungus 4 (2) 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]