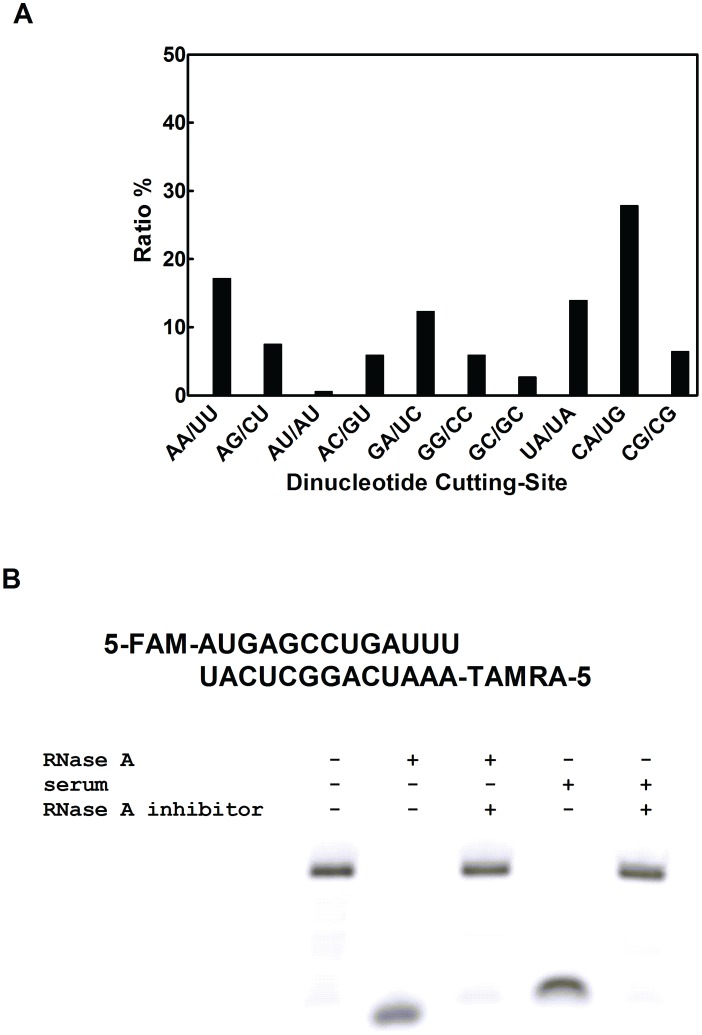
Figure 1. Cleavage preferences of duplex RNAs.
A) Degradation of an 1860 bp long RNA duplex was performed in RNase A. Degradation fragments were extracted and sequenced. By aligning the degradation fragments with the original RNA sequence, cleavage sites were identified and presented in terms of the ratio for each possible dinucleotide site [31]. B) Duplex RNA degradation assay. Dual-labeled duplex RNA was separately incubated with RNase A, the mixture of RNase A and RNase A inhibitor, human serum, the mixture of human serum and RNase A inhibitor. Samples were collected and run in a denatured PAGE gel.