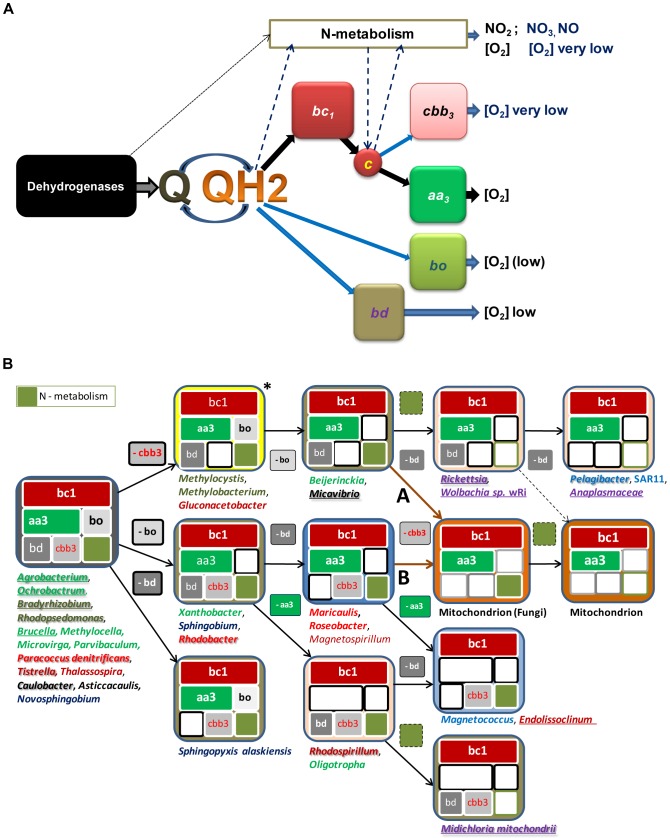
Figure 1. Bioenergetic systems of bacteria and mitochondria.
A -Terminal respiratory chain of bacteria. 11. Various bioenergetic systems - membrane redox complexes identified by their common name and different colours - carry out the oxidation of quinols (QH2) reduced by dehydrogenases. Besides oxygen (O2), nitrogen compounds can function as electron acceptors for the oxidation of dehydrogenases (dotted arrow), quinols and cytochrome c (dashed dark blue arrows), in reactions catalysed by enzyme complexes such as Nrf nitrite reductase [32], which are included within the N-metabolism system. Thick black arrows indicate electron transport in aerobic bacteria and mitochondria. Blue arrows indicate other electron transport pathways of facultatively anaerobic bacteria. B - Pathways of mitochondrial bioenergetic evolution. The bioenergetic systems illustrated in A are indicated by the coloured modules (with size proportional to their bioenergetic output) within the boxes representing the bioenergetic subset of each organism or organelle. Mitochondria of fungi and heterokont microorganisms differ from those of other eukaryotes for the presence of elements of N-metabolism. Representative taxa with fully sequenced genome are listed beneath each subset. The pathways of mitochondrial evolution are deduced by connecting these subsets with stepwise loss of a single bioenergetic system. Microorganisms underlined are symbionts or pathogens. Bacteria in embossed typeface have been proposed as ancestors or relatives of mitochondria (see Table S1 in File S1 for specific references). Dark brown arrows A and B indicate the pathways leading to fungal mitochondria. The pathway between the Rickettsia subset and that of mitochondria (dashed arrow) can be discounted, since the symbiotic event occurred only once [1], [5], [6], [10], [48]. * indicates the subset from which other pathways depart (Figure S1 in File S1).