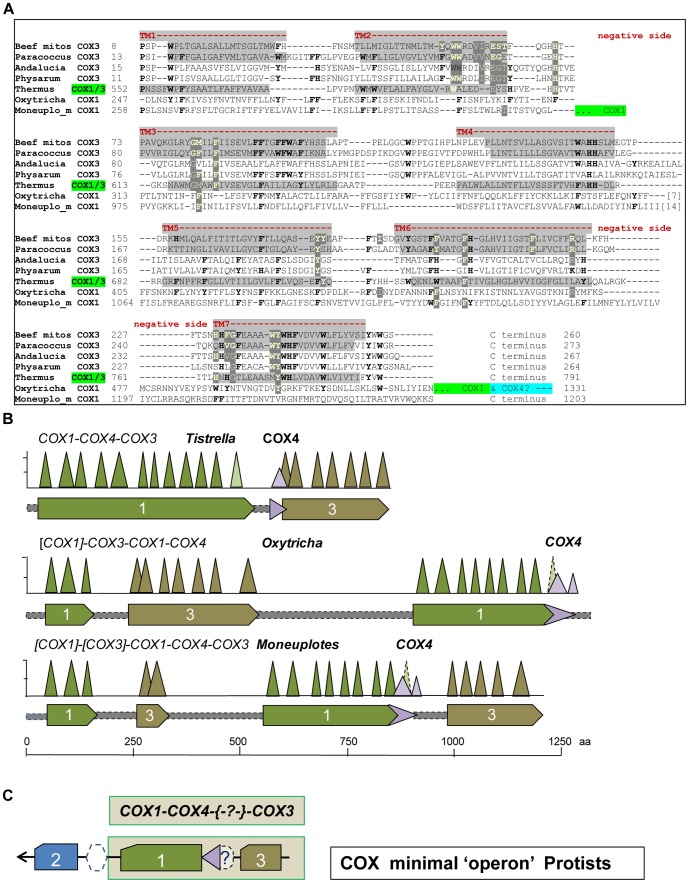
Figure 4. Analysis of the molecular architecture of COX3 in bacteria and protists. A – Alignment of bacterial and mitochondrial COX3 proteins.
A set of aligned COX3 sequences from bacteria and protists was initially obtained from the DELTABLAST option of multiple alignment and subsequently implemented manually following data available from the structure of beef [59], [60], Paracoccus [61] and Thermus [54] aa3 oxidase. Residues that bind phospholipids with either H or π bonds [60] are in yellow character and highlighted in dark grey, while those conserved are in bold character. Light grey areas indicate transmembrane helices (TM). B – Graphical representation of COX 1-3 fused proteins. The hydrophobic peaks in the hydropathy profile of the proteins, which was obtained using the program WHAT [81] with a fixed scanning window of 19 residues, is represented by the sharp triangles, that are commensurated to the peak height (maximum in the hydrophobicity profile) and width of the predicted TM [81], which closely correspond to those observed in 3D-structures [47], [54], [61]. C – Deduced sequence of the “minimal” COX operon of protists. The arrangement of COX genes essentially corresponds to the core sequence of a COX operons of type a (cf. Fig. 3) but in the reverse order of transcription. Dashed symbol represents a protein that may intermix with other COX subunits such as a COX4-like (Fig. S2 in File S1).