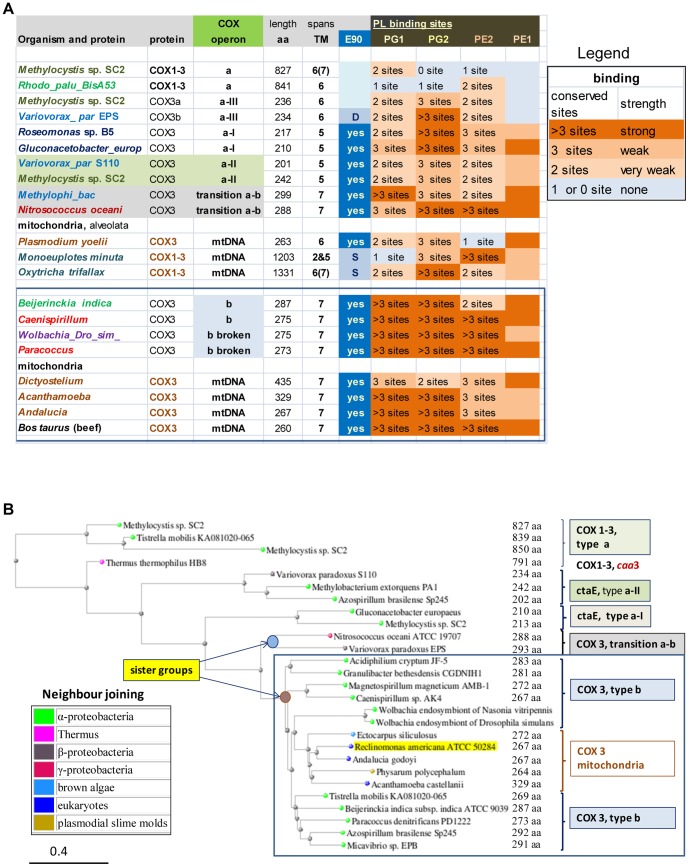
Figure 5. Structure-function features of COX3 gradually evolved from bacteria to mitochondria.
A – Heatmap for the strength of phospholipid binding by COX3 proteins.
The table summarises the molecular features of PL-binding sites (residues) in aligned COX 3 proteins (Table S4 in File S1); it is colour mapped according to the number of conserved sites to represent the increasing PL-binding strength along bacterial and mitochondrial protein sequences, as indicated by the legend on the right of the table. PL-binding is considered weak when less than 3 sites are conserved for each PL, the nomenclature of which is taken from Ref. [60]. PE, phosphatidyl-ethanolamine; PG, phosphatidyl-glycerol. The list includes conserved amino acids corresponding to E90 in beef COX3, which lies near bound PL modulating oxygen entry into the catalytic site of the oxidase [60]. Abbreviations for organisms are: Rhodo_palu_BisA53, R. palustris BisA53; Variovorax_ par, Variovorax paradoxus; Methylophi_bac, Methylophilales bacterium HTCC2181; Wolbachia_Dro_sim_, Wolbachia endosymbiont of Drosophila simulans. B - Representative distance tree of COX 3 proteins. The tree was obtained as described in the legend of Fig. 3B, using as a query the C-terminal region of the COX1-3 protein from R. palustris BisA53 (Accession: YP_782773, residues 550 to 841) that aligns with bacterial and mitochondrial COX3 (Fig. 3B Fig. 4A). The group containing bacterial proteins from COX operon type b and their mitochondrial homologues is enclosed in a blue square as in Fig. 3B.