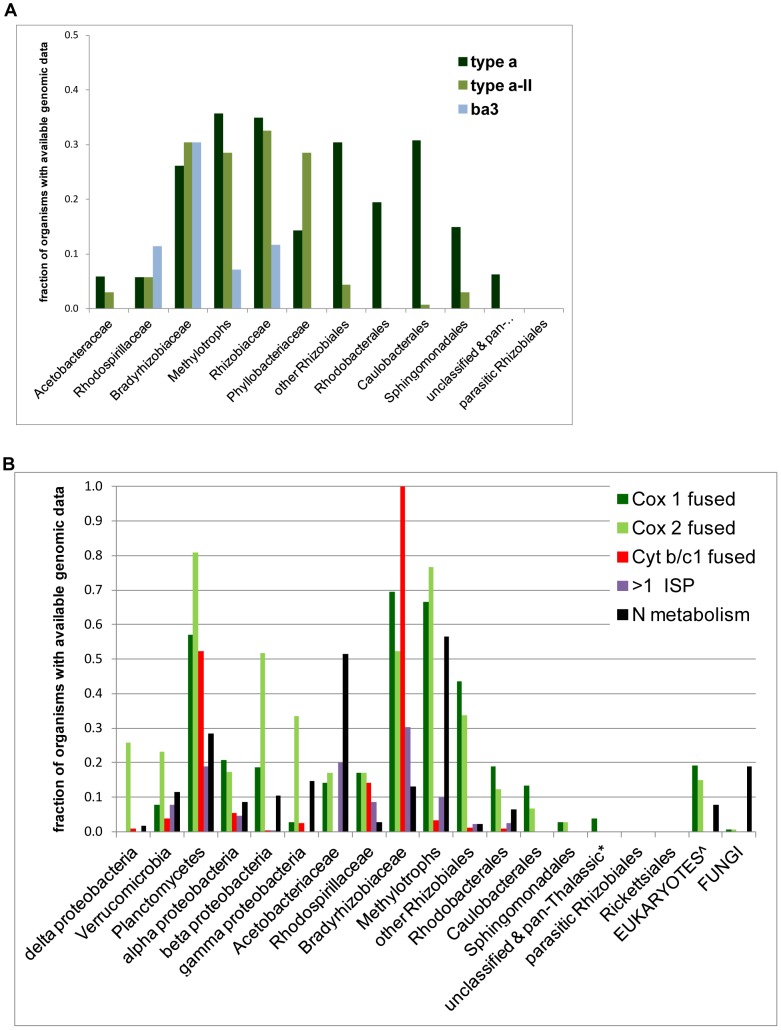
Figure 6. Taxonomic distribution of bioenergetic systems in bacteria.
A – Distribution of COX operon types in major families of α-proteobacteria.
The frequency of each type of COX operon was normalised to the number of α-proteobacterial organisms with genomic data that are currently available (from NCBI resources http://www.ncbi.nlm.nih.gov/taxonomy/- accessed 14 March 2014) [50]. See Table S2 in File S1 for a detailed list of the taxonomic distribution of diverse COX operon types. The definition ‘pan-Thalassic’collects together organisms of the SAR clade with Magnetococcus, Pelagibacter and Micavibrio. B. -Distribution of fused proteins and N-metabolism elements along diverse bacterial lineages. Fused proteins were identified with the combined resourses of NCBI and the Protein Family website (PFAM 27.0 - http://pfam.sanger.ac.uk/ [52]). Multiple forms of ISP were counted as >1 ISP. Taxa are arranged according to their approximate phylogenetic position considering also metabolic features (cf. Refs [5], [31]). For each group, the frequency is normalized as in A. Eukaryotes (∧) include amoebozoa, ciliates and heterokonts. N-metabolism encompasses: methane monooxygenase, ammonia monooxygenase, nitrite oxidoreductase, Nirf nitrite reductase and its homologues in COX operon type a-I (Fig. 3A), ammonia oxidation and anaerobic ammonia fermentation [30], [32].