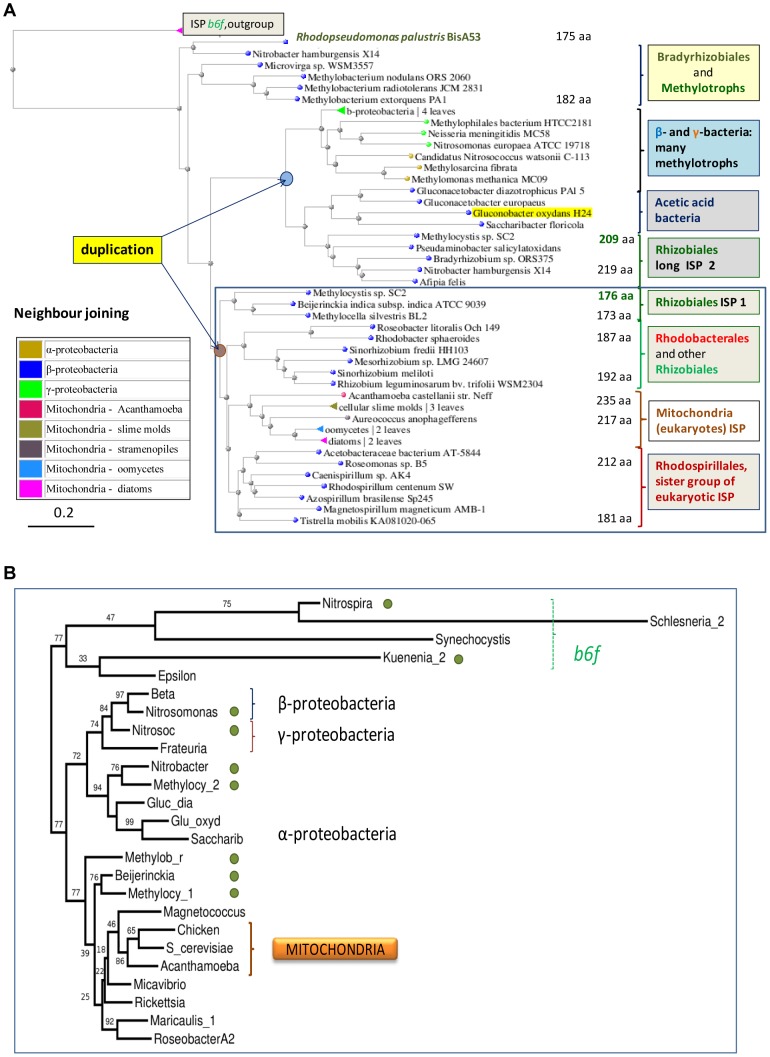
Figure 8. Phylogenetic relationships between diverse forms of ISP.
A – Distance tree encompassing proteobacteria and mitochondria. The tree was obtained as described in the legend of Fig. 3B using the alignment of Fig. 7A and two ISP proteins from the b6f complex as outgroup (top). The group containing bacterial ISP1 proteins together with their mitochondrial homologes is enclosed in the blue square to highlight a likely ancestral duplication separating it from the group with ISP2. B – Long distance phylogenetic relationships of bacterial ISP. The phylogenetic tree (maximal likelyhood method) of ISP proteins was computed from the structure-based alignments in Fig. S4 in File S1. Th small green circle indicates ancient nitrogen or methylotrophic metabolism [29]–[32] (Fig. 6B). The dashed green bracket indicates the paralogue proteins belonging to the b6f complex. Other brackets indicate proteobacterial subdivisions and mitochondria as in A. Note how the bootstrap values are much lower within the bottom branch containing mitochondrial ISP than in the upper branch containing ISP2.