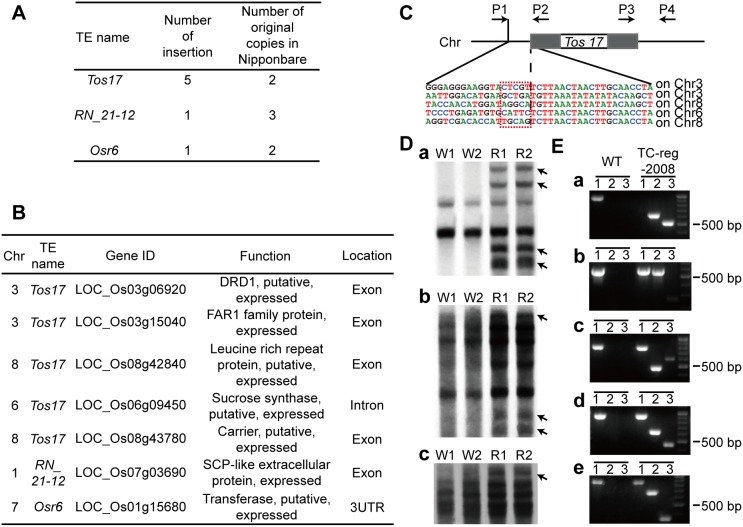
Figure 3. Detection and validation of TE mobility.
(A) Three active retrotransposons and seven insertions detected by paired-end mapping based on genome re-sequencing. (B) Insertion positions of all the activated retrotransposons and the function analysis of affected genes in TC-reg-2008. (C) Location of primers designed for locus specific-PCR and the junctions of the Tos17 insertion sites detected by Sanger-sequencing. P1/P4, P1/P2 and P3/P4 were three pairs of primers for amplifying the insertions of Tos17. The grey rectangles stand for LTRs of Tos17. The target site duplications of Tos17 were boxed in red based on Sanger-sequencing. (D) Independent validation of the mobilization events by Southern blotting. a. Tos17 probe, b. RN_21–12 probe, c. Osr6 probe. W1 and W2 stand for individual plants of wild type (WT), and R1 and R2 stand for individual plants of TC-reg-2008. (E) Five insertions by retrotransposon Tos17 were validated by locus-specific PCR. Lanes 1, 2 and 3 represent the PCR amplicons by primers P1/P4, P1/P2 and P3/P4, respectively.