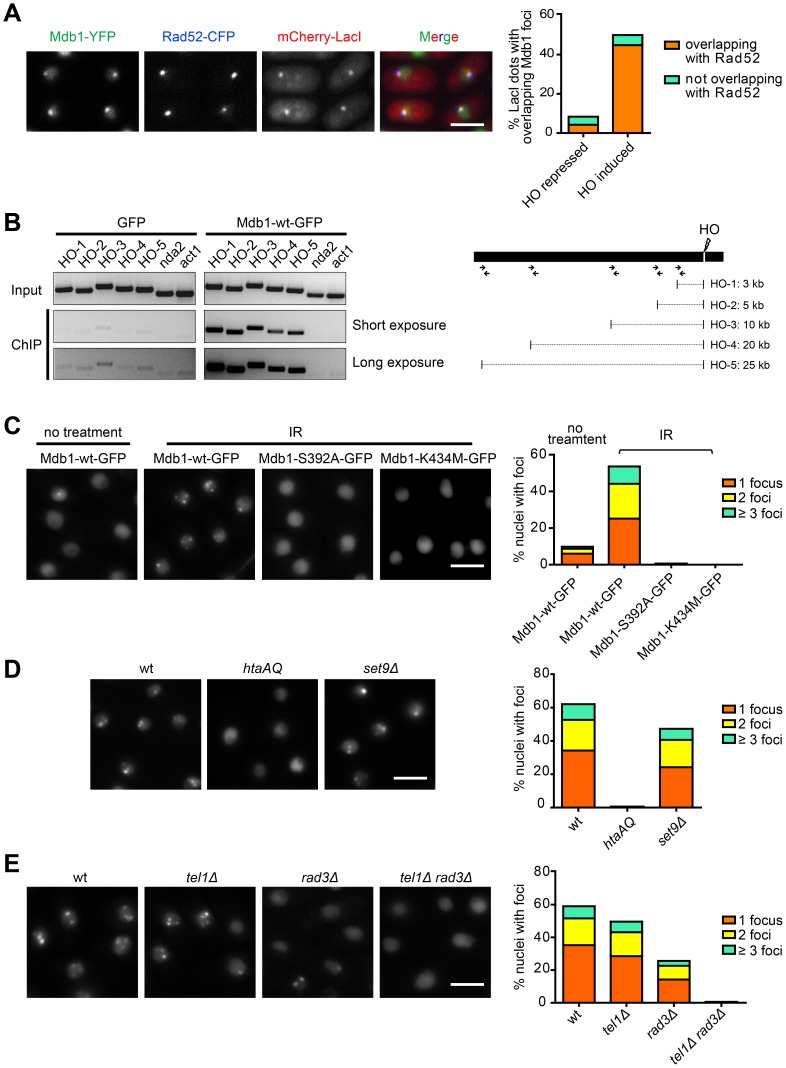
Figure 2. Mdb1 relocalizes to DSBs in a γH2A-dependent manner.
(A) Mdb1-YFP forms nuclear foci at HO-induced DSBs. For HO endonuclease induction, cells were shifted to a thiamine-free medium for 12 h before imaging. The strain used was DY389. Bar, 5 µm. (B) Chromatin immunoprecipitation (ChIP) analysis using anti-GFP antibody showed that Mdb1-GFP, but not GFP alone, is recruited to chromatin regions adjacent to the HO-induced DSB. PCR primers used for amplifying DNA regions near the HO cleavage site are depicted in the schematic on the right. Primers amplifying the genes nda2 and act1 were used as negative controls. The strains used were DY16261 and DY16263. (C) The phospho-binding residues are required for Mdb1 IRIF formation. Mdb1 IRIF were imaged and quantified before and after exposure to 36 Gy of IR. To quantify the levels of foci-containing nuclei, about 200 nuclei were examined for each sample. Strains used were DY15603, DY15604, and DY15605. Bar, 5 µm. (D) Mdb1 IRIF formation requires γ-H2A but not H4K20me. Cells were treated with 36 Gy of IR and analyzed as in (B). Strains used were DY15603, DY15606, and DY15607. Bar, 5 µm. (E) Mdb1 IRIF formation depends on Tel1 and Rad3. Cells were treated with 36 Gy of IR and analyzed as in (B). Strains used were DY15631, DY15608, DY15609, and DY15610. Bar, 5 µm.