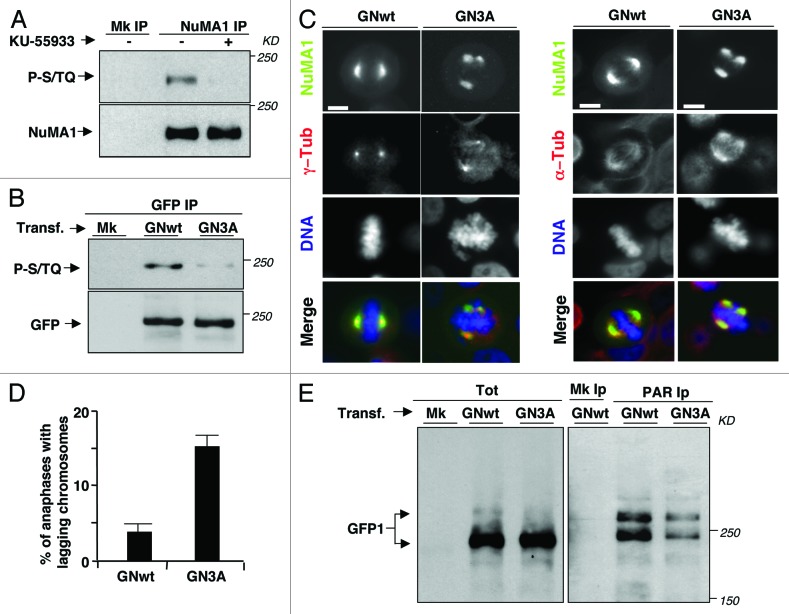
Figure 5. NuMA1 phosphorylations affecting spindle assembly and PARylation. (A) HeLa cells were treated as described in Figure 2A; mock (Mk) or NuMA1 IPs probed for phospho-S/TQ sites (P-S/TQ; top panel), NuMA1 (bottom panel). (B) HeLa cells were either mock (Mk) or transfected with a GFP-NuMA1-wild type (GNwt) or with GFP-NuMA1–3-ser-to-ala mutant (GN3A) fusion protein expression vectors 24 h prior to a 14 h nocodazole treatment. GFP IPs probed for phospho-S/TQ sites (P-S/TQ; top panel) and for GFP (bottom panel). (C) HeLa cells were transfected with GNwt or GN3A expression vectors. Twenty-four hours post-tranfections, cells were fixed and immunostained for (left panels) γ-tubulin (red), or (right panels) α-tubulin (red), the GFP-NuMA1 fusion proteins were visualized by autofluorescence (green) and DNA by Hoechst 33258 staining (blue). Scale bars, 10 μm. (D) Histogram: percent of anaphases with lagging chromosomes in GNwt- and GN3A-transfected HeLa cells. Error bars indicate s.d. (E) HeLa cells were either mock transfected (Mk) or transfected with GNwt or GN3A expression vectors 60 h prior to 14 h-nocodazole treatment. Detached cells were isolated, and total cytosolic extracts (Tot), mock (Mk), and anti-PAR IPs were probed with an anti-GFP antibody. In transfected cells, the faint bands recognized by the anti-GFP antibody migrating above the 250-kD marker in total lysates perhaps represent hyper-PARylation or other modifications of PARylated GFP-NuMA1, since these appear enriched in PAR IPs (especially in the GNwt-transfectants) and are never detected in non- or mock-transfected cells. The data shown are representative of 3 independent experiments per type.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.