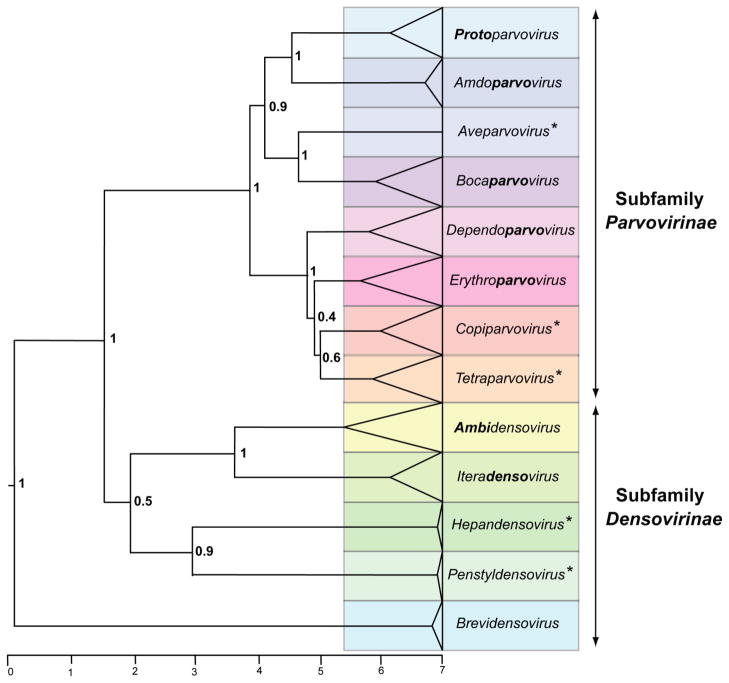
Figure 1. Phylogenetic tree showing genera in the family Parvoviridae.
Phylogenetic analysis based on the amino acid sequence of the viral replication initiator protein, NS1, which contains a conserved AAA+ helicase domain corresponding to the Parvo_NS1 Pfam domain: http://pfam.sanger.ac.uk/family/Parvo_NS1. This region was aligned by incorporating insights from structural biology using the ehmmalign application in EMBASSY [6], and sequences flanking the Pfam domain were aligned using the modification of the Needleman-Wunsch local alignment method [11] as implented in MOE-Align ( http://www.chemcomp.com). Pairwise p-distance matrices were constructed from this alignment using MEGA version 5.10 [12]. Bayesian trees were calculated over one billion iterations using BEAST [5], using a Yule model of speciation and an exponential relaxed molecular clock [4]. Trees were viewed in FigTree (part of BEAST) in ultrametric format on an arbitrary scale, midpoint-rooted, and with posterior probability scores indicated at statistically significant nodes. Bold type in genus names indicates affixes used to expand existing names. Asterisks denote the names of new genera.