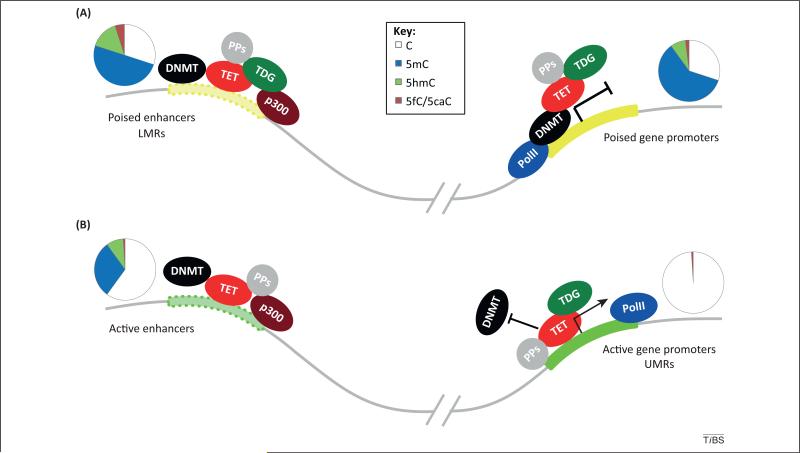
Figure 2.
Schematic representation of DNA epigenetic regulation in mouse embryonic stem cells (ESCs). TET/thymine DNA glycosylase (TDG)-mediated active DNA demethylation occurs frequently at distal regulatory elements in ESCs. (A) At poised enhancers and poised gene promoters, where 5-formylcytosine (5fC)/5-carboxylcytosine (5caC) accumulation in the absence of TDG is the most significant, dynamic methylation and demethylation equilibria likely exist to keep these regions at the poised states in order to respond to upcoming development/differentiation events. Accordingly, these 5fC/5caC-marked 5-hydroxymethylcysotine (5hmC) sites are committed demethylation sites. (B) 5hmC and 5fC/5caC are also present at active enhancers where they may have more active regulatory roles. Certain active promoter sites (unmethylated regions; UMRs) are depleted of 5mC and 5hmC but still bound by TET proteins. 5hmC and 5fC may recruit binding proteins to facilitate active demethylation. TET/TDG may interact with diverse protein partners (PPs) that affect their demethylation activities. ESC DNA methylation patterns are represented by pie charts above each gene elements. Abbreviations: DNMT, DNA methyltransferease; LMRs, low-methylated regions; PolII, RNA polymerase II.