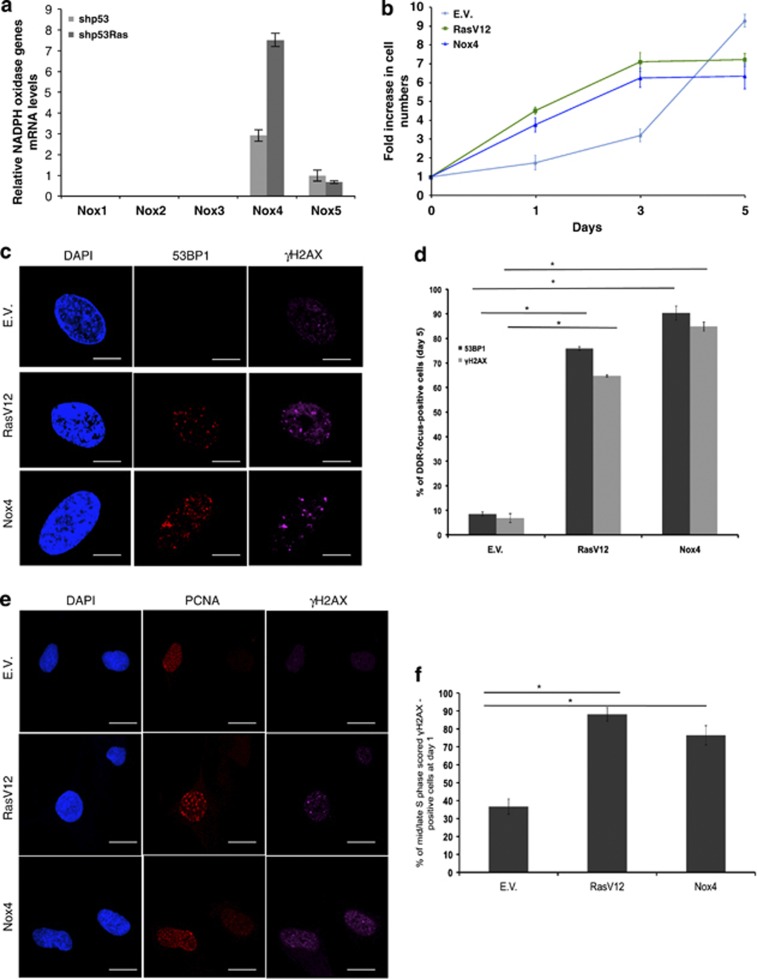
Figure 3.
Nox4 is induced by H-RasV12 and its expression is sufficient to drive hyperproliferation, DDR activation and proliferative arrest. (a) qRT-PCR analysis of known human NOX gene paralogs shows that NOX4 is induced upon RasV12 expression. (b) Nox4 overexpression in NHFs is sufficient to induce hyperproliferation and a proliferative arrest similar to RasV12 expression. Proliferation is shown as the ratio of fold increase in cell numbers compared with day 0 (initial plating day) For further details, see Materials and Methods and also Supplementary Figure S3. (c) Nox4 overexpression triggers DDR to an extent similar to RasV12 expression. Confocal images of DDR markers immunostained for 53BP1 and γH2AX in E.V., RasV12 and Nox4-transduced cells. The percentages of DDR in the form of γH2AX and 53BP1 foci of cells are compared individually among RasV12- or Nox4-expressing cells with respect to E.V. Error bars indicate S.E.M. (n≥ 3), and differences are statistically significant (*P-value < 0.01) throughout the figure where stated. Scale bar: 2.8 μm. (d) RasV12 or Nox4 expression induces similar levels of DDR as shown by quantification of DDR in the form of 53BP1 and γH2AX foci in cells at the last time point of the growth curve in 3B. (e) Confocal images of DDR markers immunostained for γH2AX in E.V., RasV12 and Nox4 lentiviral-transduced cells demonstrate that DDR activation was induced similarly in both RasV12 and Nox4 expressing cells. The mid or late S phase stage of the cell cycle was established by scoring nuclear PCNA staining patterns.66 Scale bar: 12 μm. (f) RasV12 or Nox4 expression with respect to E.V. infected NHF cells induces similar levels of DNA replication stress as shown by the quantification of γH2AX focus formation with cells having a mid or late S phase PCNA foci at day 1 of the growth curve in panel b. The percentages of DDR in the form of γH2AX foci of cells are compared individually among RasV12- or Nox4-expressing cells with respect to E.V. Error bars indicate S.E.M. (n≥ 3), and differences are statistically significant (*P-value < 0.01) throughout the figure where stated