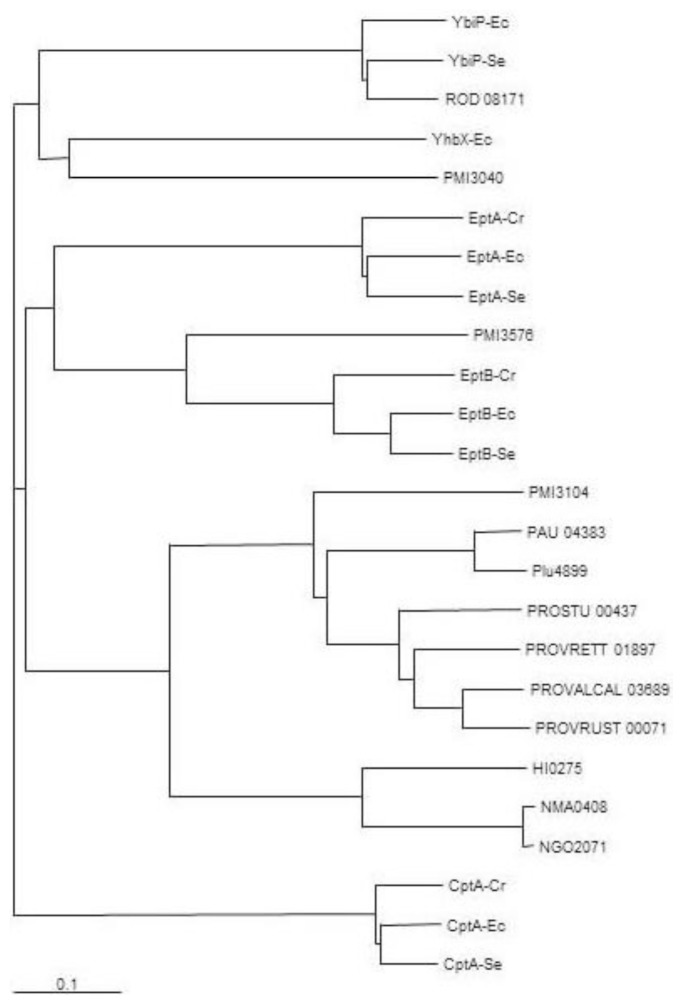
Figure 2.
Phylogenetic tree of selected known and putative PEtN transferases. Shown are proteins from P. mirabilis HI4320 (PMI3040, PMI3576, PMI3104 [eptC]), E. coli MG1655 (YbiP-Ec, YhbX-Ec, CptA-Ec, EptA-Ec, EptB-Ec), S. enterica LT2 (YbiP-Se, CptA-Se, EptA-Se, EptB-Se), Citrobacter rodentium ICC168 (ROD 08171, CptA-Cr, EptA-Cr, EptB-Cr), Photorhabdus asymbiotica asymbiotica ATCC 43949 (PAU 04383), Pho. luminescnes laumondii TT01 (Plu4899), Providencia stuartii ATCC 25827 (PROSTU 00437), Providencia rettgeri DSM 1131 (PROVRETT 01897), Providencia alcalifaciens DSM 30120 (PROVALCAL 03689), Providencia rustigianii DSM 4541 (PROVRUST 00071), Haemophilus influenzae Rd KW20 (HI0275), Neisseria meningitidis Z2491 (NMA0408), and N. gonorrhoeae FA 1090 (NGO2071). Note: The separation and the size of the lanes are relative to the similarity degree according to the program used (the scale bar indicates an evolutionary distance of 0.1 amino acid substitutions per position).