FIG. 2.
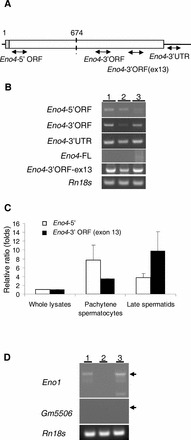
A–C) Expression of Eno1 transcripts and variants. A) The location of primers specific for each of the Eno4 transcripts used in the PCR assays. B) Conventional RT-PCR results with specific primer pairs detecting Eno4 transcript and variants in testes and isolated germ cells. Lanes contained total RNA from testes (lane 1), StaPut-isolated pachytene spermatocytes (lane 2), and StaPut-isolated round spermatids (lane 3). Thirty-five amplification cycles were used for the Eno4 transcript and variants, and the annealing temperature used was 63°C. C) Expression of Eno4 transcripts in pachytene spermatocytes and late spermatids isolated by LCM. Primers specific for each of the Eno4 transcripts was used to assay by qPCR their expression in whole lysates, pachytene spermatocyte, and late spermatids; Eno4-5′, white bars; Eno4-3′ORF (exon 13), black bars. Expression levels were determined as described in Materials and Methods and shown here as ratios (folds) relative to the level on whole lysate, with that level set at one. Data are expressed as means ± SEM. D) Expression of Gm5506 and Eno1 transcripts in mouse testes. Gene specific primers were used to detect Eno1 and Gm5506 using cDNA from testes and isolated germ cells by conventional RT-PCR. The amplification cycles of PCR for Eno1 and Gm5506 were 40 or 50, respectively. The annealing temperature used for Eno1 or Gm5506 was 60°C. Lanes contained total RNA from testes (lane 1), StaPut-isolated pachytene spermatocytes (lane 2), and Sta-Put-isolated round spermatids (lane 3). 18S ribosomal RNA (Rn18s) was used as a control.
